Figure 4.
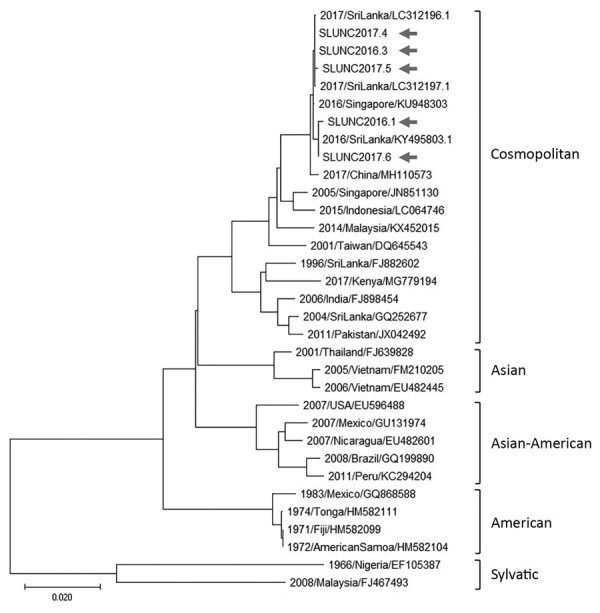
Phylogenetic tree for 5 dengue virus 2 (DENV-2) isolates from late 2016 and 2017 dengue epidemic (arrows), Sri Lanka, and reference DENV-2 strains. The tree is based on a 1,485-nt fragment that encodes the envelope protein. Classification and naming of DENV-2 genotypes are based on (16). The evolutionary history was inferred using the neighbor-joining method (17). The optimal tree with the sum of branch length = 0.44012906 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the maximum composite likelihood method (18) and are in the units of the number of base substitutions per site. This analysis involved 33-nt sequences. All ambiguous positions were removed for each sequence pair (pairwise deletion option). The final dataset comprised 1,485 positions. Evolutionary analyses were conducted in MEGA X (12). Scale bar indicates nucleotide substitutions per site.
