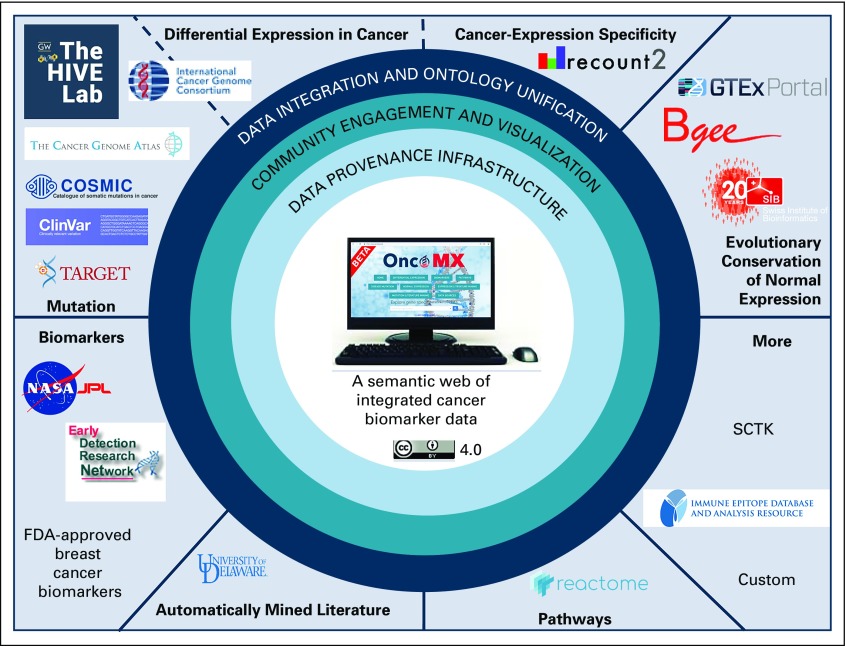
FIG 1.
OncoMX overview. External segments: Data for cancer mutation, cancer expression, healthy expression, literature mining for both cancer mutation and expression, cancer biomarkers, relevant ontologies, functional annotations, and pathways were initially harvested from a number of publicly available resources, such as The Cancer Genome Atlas, International Cancer Genome Consortium, ClinVar, COSMIC, UniProt, and others, and further processed or analyzed by The George Washington University for BioMuta and BioXpress; SIB (Swiss Institute of Bioinformatics) for Bgee (RNA sequencing–derived healthy expression calls and ranks for human and mouse); the University of Delaware for literature mining through DiMeX and DEXTER; and the Early Detection Research Network for biomarkers. Outer ring: Data sets were integrated and unified through Cancer Disease Ontology slim terms for disease names and Uberon Anatomical Entity terms for tissue and physiologic location. Middle ring: Feedback was solicited through a multipronged approach involving use case collection at poster sessions and two formal workshops. With user community guidance, a series of data views and graphic visualizations were devised to aid end users in the exploration and interpretation of biomarker evidence. Inner ring: Data sets were refined and documented with provenance details following the BioCompute Object model for data provenance capture and hosted through the data.oncomx.org data site. Inner circle: The product of these efforts is the current OncoMX Web portal, a semantic web of integrated cancer biomarker data that is readily accessible and licensed under a Creative Commons Attribution 4.0 International License. FDA, US Food and Drug Administration; SCTK, Single-Cell Toolkit.40