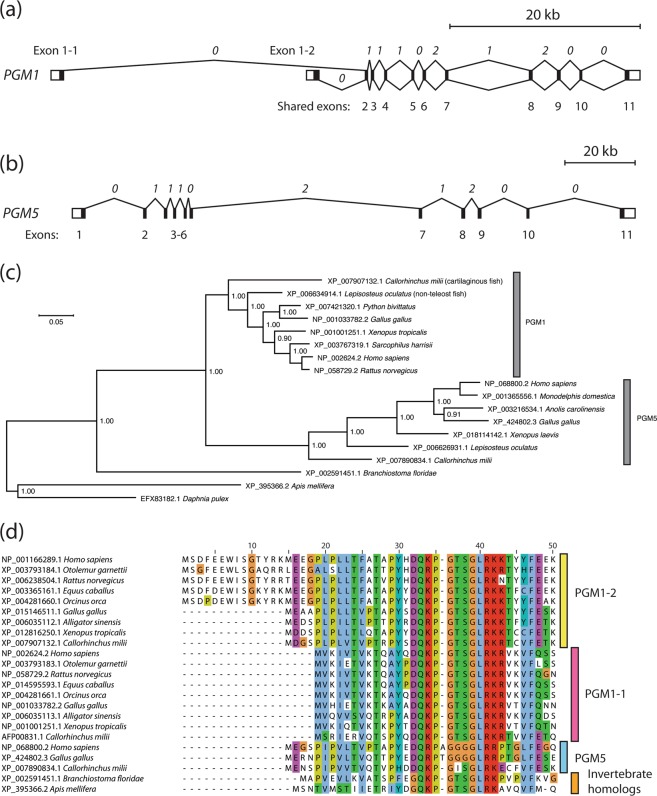
Figure 1.
PGM1 sequence evolution. (a) Gene structure of human PGM1 with two alternative transcription start sites in exons 1-1 and 1-2 giving rise to the alternative transcripts translated into PGM1 isoforms 1 (PGM1-1, splicing shown above the exons) and 2 (PGM1-2, splicing shown below exons). 5′ untranslated regions (5′ UTRs) are shown in exons 1-1 and 1-2, and 3’ UTR in exon 11, as white boxes. Intron phases are shown (in italics) above the introns, defined as the position of the intron within a codon, with introns with phases 0, 1, and, 2 located before the first base, after the first base, or after the second base, respectively. The gene is found at chromosome 1p31.3 and has a length of approximately 70,000 base pairs (see also scale bar above gene structure). The exon lengths are not shown at the correct scale. (b) Gene structure of human PGM1 paralog PGM5 with 5′ and 3′ UTRs shown as white boxes in exons 1 and 11, respectively. Intron phases, conserved and identical to PGM1, are shown (in italics) above the introns. The gene is located at chromosome 9p21.11 with a length of approximately 175 kb. (c) The PGM1 and PGM5 paralogs are the result of a gene duplication event in the common ancestor of the Gnathostomata, of jawed vertebrates. The Bayesian inference tree is based on the alignment of the protein sequences (segment corresponding to exons 2 to 11 in human PGM1, WAG + Γ model with four rate categories). The phylogram is shown with estimated branch lengths proportional to the number of substitutions at each site, as indicated by the scale bar. For each node, Bayesian posterior probabilities are shown. The arthropod clade with the crustacean D. pulex and the honey bee (A. mellifera) was set as outgroup in order to root the tree. (d) Multiple sequence alignment of the N-termini of isoforms PGM1-1 and PGM1-2 from human, northern greater galago, rat, horse, orca, chicken, alligator, a frog and the Australian ghostshark, a cartilaginous fish (binomial names of the species, in the same order, is given in the panel), together with three vertebrate PGM5 sequences and homologs from two invertebrates, the amphioxus and honey bee. Residue numbering for human PGM1-2 is shown above the alignment.