Figure 2.
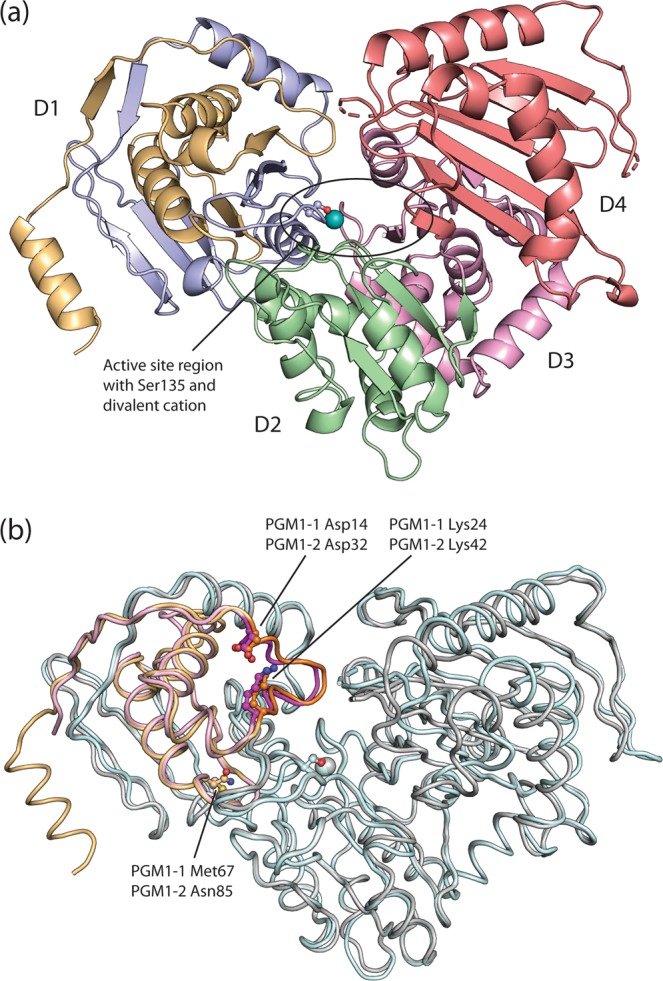
Structure of human PGM1 isoform 2. (a) PGM1-2 has four structural domains, D1 to D4, arranged in an overall “heart shape”. Domains D2 to D4 are shown in green (residues 210-322), pink (residues 323-439), and red (residues 440-580), respectively. The segment of D1 encoded by isoform specific exon 1-2 is shown in light orange (N-terminus to residue 100), while the rest of D1 (residues 101 to 209), encoded by exons 2, 3, and partially by exon 4, is colored blue. The active site region, with contributions from all four domains, has a dephosphorylated Ser135 (ball-and-stick representation) and binds a divalent cation (teal sphere). It is located in the central large cleft. (b) Structural alignment of the recently solved 3D structure of PGM1-115 (PDB identifier 5EPC, chain A) and PGM1-2 (this work) reveals that the two isoforms are highly similar. The exon 1-1 encoded segment of PGM1-1 D1 is colored pink, while the rest of the protein is shown in pale cyan. PGM1-2 is shown in grey, with the isoform specific exon 1-2 encoded segment in orange. Two isoform specific, paralogous loops, Asp14 to Lys24 in PGM1-1 and Asp32 to Lys42 in PGM1-2, are shown with darker shades of pink and orange, respectively. The loops have identical sequences, very similar structures and are the only parts of the exon1-1/exon1-2 encoded segments that are relatively close to the active site. PGM1-1 Met67 and PGM1-2 Asn85 is the pair of paralogous non-identical residues that are closest to the active site (>10 Å), strongly suggesting that PGM1-1 and PGM1-2 have identical active sites. The figure is rendered as a Cα trace.
