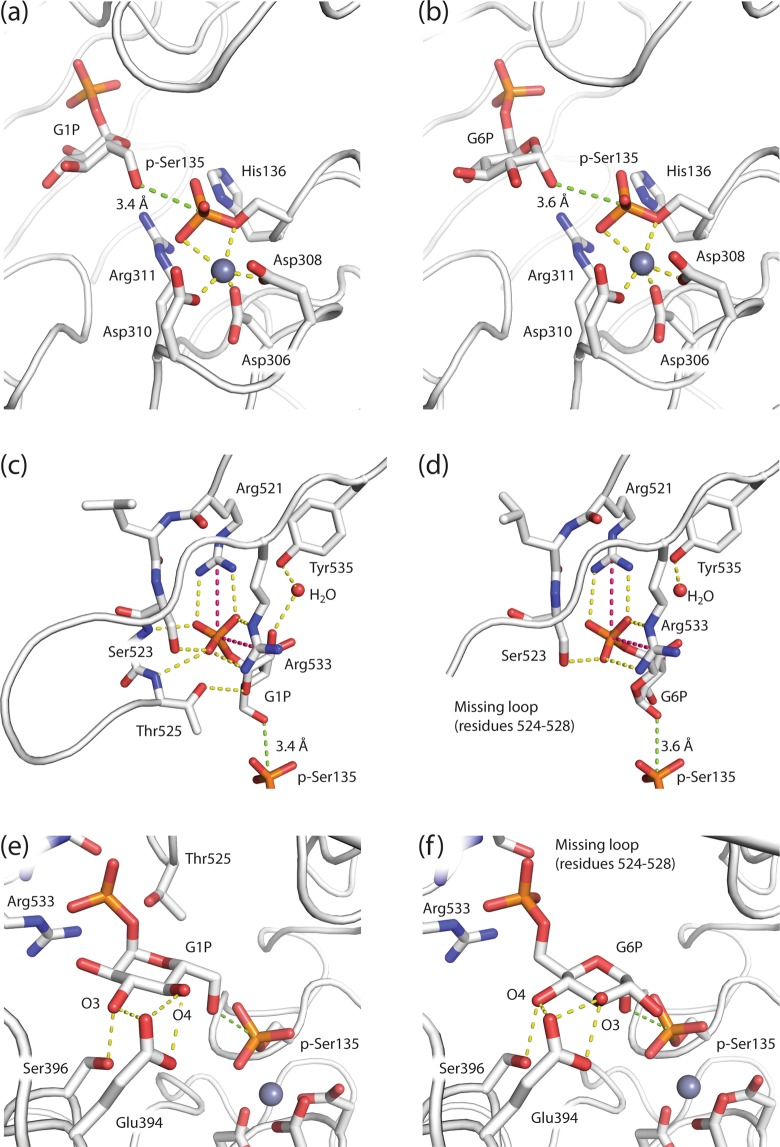
Figure 5.
Comparison of the PGM1 substrate and product complex active sites. (a) Substrate (G1P) and (b) product (G6P) are located with O6 and O1, respectively, roughly 3.5 Å (green dotted line) from the P atom of the phosphoserine (p-Ser135) of the PGM1 active site. The divalent cation (grey sphere) is complexed by three Asp residues of the metal-binding loop in D2 and the p-Ser group (yellow dotted lines). Both substrate (c) and product (d) are being anchored in the active site by strong, bi-dentate interactions (pink and yellow dotted lines) to Arg521 and Arg533 in the phosphate-binding loop of D4. Due to lacking electron density, residues 524 to 528 are not visible in the PGM1-2:G6P structure. With the hexose phosphate group anchored by the phosphate-binding loop, the residues Glu394 and Ser396 of the sugar-binding loop in D3 recognizes the hydroxyls in their equatorial positions at C3 and C4 in substrate G1P (e). The same two residues, in essentially identical conformations, contact the two hydroxyls, but in swapped positions, in the product complex (f).