Abstract
The purpose of this study was to assess the value of metagenomic next-generation sequencing (mNGS) of bronchoalveolar lavage fluid (BALF) for the diagnosis of severe respiratory diseases based on interpretation of sequencing results. BALF samples were harvested and used for mNGS as well as microbiological detection. Infectious bacteria or fungi were defined according to relative abundance and number of unique reads. We performed mNGS on 35 BALF samples from 32 patients. The positive rate reached 100% in the mNGS analysis of nine immunocompromised patients. Compared with the culture method, mNGS had a diagnostic sensitivity of 88.89% and a specificity of 74.07% with an agreement rate of 77.78% between these two methods. Compared with the smear method and PCR, mNGS had a diagnostic sensitivity of 77.78% and a specificity of 70.00%. In 13 cases, detection results were positive by mNGS but negative by culture/smear and PCR. The mNGS findings in 11/32 (34.4%) cases led to changes in treatment strategies. Linear regression analysis showed that diversity was significantly correlated with interval between disease onset and sampling. Dynamic changes in reads could indirectly reflect therapeutic effectiveness. BALF mNGS improves sensitivity of pathogen detection and provides guidance in clinical practice. Potential pathogens can be identified based on relative abundance and number of unique reads.
Electronic supplementary material
The online version of this article (10.1007/s10096-019-03734-5) contains supplementary material, which is available to authorized users.
Keywords: Metagenomic next-generation sequencing (mNGS), Bronchoalveolar lavage fluid (BALF), Respiratory failure, Clinical diagnosis
Introduction
The rational use of antibiotics is extremely important for immunocompromised and/or critically ill patients, for whom early aetiological diagnosis is necessary. However, traditional culture methods are time consuming and have low rates of positive detection. With the development of molecular biology, the value of whole genome–based next-generation sequencing (NGS) has gradually been recognized, especially for the detection of rare, atypical, or slow-growing microbes. NGS is mainly used in the clinical setting for assessment of sterile body fluids, including cerebrospinal fluid, blood, and joint effusion [1–3]. For non-sterile body fluids, such as sputum and bronchoalveolar lavage fluid (BALF), the applications of NGS are quite limited. In addition to human genome interference and other common problems, the interpretation of NGS results (i.e. distinguishing colonization vs. infection) remains problematic. In recent years, the feasibility of metagenomic next-generation sequencing (mNGS) in the aetiological detection and identification of respiratory tract infections has been demonstrated [4]. In the current study, we summarized the mNGS results for 35 BALF samples from 32 patients and attempted to verify the value of mNGS in the diagnosis of severe respiratory diseases based on the interpretation of sequencing results.
Materials and methods
Patients and collection of BALF samples
Patients admitted to our respiratory intensive care unit (RICU) due to respiratory failure provided written informed consent to undergo bronchoscopy and mNGS between September 2017 and October 2018; they were then examined via bedside bronchoscopy by experienced physicians. The safety of bronchoalveolar lavage (BAL) was enhanced by following a standard safety protocol [5].
BALF samples were harvested, of which 5 mL of the specimen was placed in a sterile sputum container, stored at − 20°C, and then sent to BGI-Huada Genomics Institute (Shenzhen, China) for detection. The remaining specimens were sent to our microbiological laboratory for bacterial and fungal smear and culture, Pneumocystis jirovecii (PC) smear (Grocott methenamine staining), acid-fast stain, Xpert MTB/RIF detection of DNA sequences specific for Mycobacterium tuberculosis and rifampicin resistance by PCR, and real-time PCR for cytomegalovirus (CMV), influenza A/B virus, PC, Mycobacterium tuberculosis, Mycoplasma spp., and Chlamydia spp.
NGS procedure for BALF samples
The NGS procedure for BALF samples includes nucleic acid extraction, library construction, sequencing, and information analysis. Additional data are given in Online Resource 1.
Definitions
SDSMRN, number of unique reads: the number of unique reads of standardized species
Relative abundance: proportion of a detected microbe at a specific taxonomic level among a specific microbial group in the entire sample population
Coverage: percentage of the length of the nucleic acid sequence detected over the total length of the genome of the microbe
Infectious pathogens were defined as those meeting either of the following conditions: (a) > 30% relative abundance at the genus level, regardless of culture or smear result; (b) culture and mNGS identified same microbe, and number of unique reads was ≥ 50 from a single species. Microbes were considered new potential pathogens if mNGS alone identified the microbe [3, 6]. These were based on strict clinical criteria, combined with multiple-clinician adjudication, to rigorously discriminate infection from colonization and contamination.
The definitions of ARDS and immunocompromised subjects are presented in the literatures [7, 8].
Statistical analysis
Continuous variables are presented as the mean ± standard deviation. The paired McNemar chi-square test was used to compare the diagnostic efficiency of mNGS vs. conventional smear or culture methods. Linear regression analysis was used to assess the relationships of BALF aetiology with some factors. A two-tailed P value of 0.05 was considered to indicate statistical significance. All statistical analyses were performed using SPSS 22.0 (IBM Corp., Armonk, NY, USA).
Results
Thirty-two patients with respiratory failure were admitted to the RICU, among whom 21 were men; the mean age was 54.51 ± 15.41 years. Nine immunocompromised patients were hospitalized due to pneumonia (Table 1).
Table 1.
Demographic information
| Characteristics | Patients, n (%) | |
|---|---|---|
| Age (years) | 54.51 ± 15.41 | |
| Male | 21 (65.63) | |
| Smoking | Never | 20 (64.52) |
| Ex | 8 (25.00) | |
| Current | 4 (12.9) | |
| COPD | 2 | |
| Other chronic pulmonary diseases | 2 | |
| Immunocompromised | 9 | |
Based on definitions of infection, common bacterial colonization was ruled out [9–11]. Online Resource 2 lists BALF mNGS results for 35 cases in comparison with conventional method results. PCP was detected by the smear or nucleic acid method except in two immunocompromised patients (no. 14 and no. 24); in contrast, positive rate reached 100% in mNGS analysis [12, 13].
Compared with the culture method, mNGS had a diagnostic sensitivity of 88.89% and a specificity of 74.07% with an agreement rate of 77.78% between these two methods. Compared with the smear method and PCR, mNGS had a diagnostic sensitivity of 77.78% and a specificity of 70.00% with an agreement rate of 73.68% between these two methods. The overall diagnostic agreement rate was 75.68%, and the sensitivity was 81.48% (Table 2). In 13 cases, the detection results were positive by mNGS but negative by culture/smear and PCR. mNGS did not increase the positive rate of diagnosis compared with culture or smear/PCR (P = 0.070 and 0.754, respectively).
Table 2.
Diagnostic performance of mNGS compared with that of conventional laboratory-based diagnostic methods
| mNGS | Culture | Total | Smear/PCR | Total | ||
|---|---|---|---|---|---|---|
| + | − | + | − | |||
| + | 8 | 7 | 15 | 14 | 6 | 20 |
| − | 1 | 20 | 21 | 4 | 14 | 18 |
| Total | 9 | 27 | 36 | 18 | 20 | 38 |
The mNGS findings in eleven cases led to a change in treatment strategies, including four positive mNGS findings (Chlamydia psittaci in two cases and Nocardia sp. in two cases). For patient no. 20 with Acinetobacter baumannii infection and no. 25 with Aspergillus niger infection, mNGS provided positive identification of the pathogen earlier than was permitted by the culture method, leading to prompt changes in the anti-infection treatment. For patient no. 10 with negative laboratory and mNGS outcomes combined with other autoimmune indicators, dermatomyositis was considered after consultation with the rheumatology department, and 160 mg methylprednisolone was given for initial therapy. The results of both mNGS and conventional methods were negative for patient no. 27; due to the combination of these findings with a lung CT result, organized pneumonia after infection was considered, and 80 mg of methylprednisolone was initiated (Table 3).
Table 3.
The mNGS findings that led to changes in treatment strategy for 11 patients
| Patient no. | mNGS-based diagnosis | Changes in treatment strategies |
|---|---|---|
| 2 | Chlamydia sp. pneumonia | Discontinuation of Tamiflu and Sulperazon |
| 4 | Chlamydia sp. pneumonia | Discontinuation of Tamiflu and Sulperazon |
| 9 | Severe pneumonia | Discontinuation of moxifloxacin and Sulperazon |
| 10 | Dermatomyositis | Methylprednisolone (160 mg) initiated |
| 11 | Human adenovirus pneumonia | Added ganciclovir and discontinued Sulperazon |
| 13 | HSV1 pneumonia? | Meropenem downgraded to Tazocin |
| 17 | Bacterial infection | Meropenem changed to tigecycline and Sulperazon |
| 21 |
Nocardia sp. pneumonia PCP CMV pneumonia |
Moxifloxacin was replaced by amikacin, and antituberculosis drugs were discontinued |
| 25 | Aspergillus sp. pneumonia | Added voriconazole and discontinued moxifloxacin |
| 27 | Organized pneumonia | Methylprednisolone (80 mg) initiated |
| 32 |
Nocardia sp. pneumonia PCP CMV pneumonia |
Sulperazon changed to imipenem and cilastatin sodium |
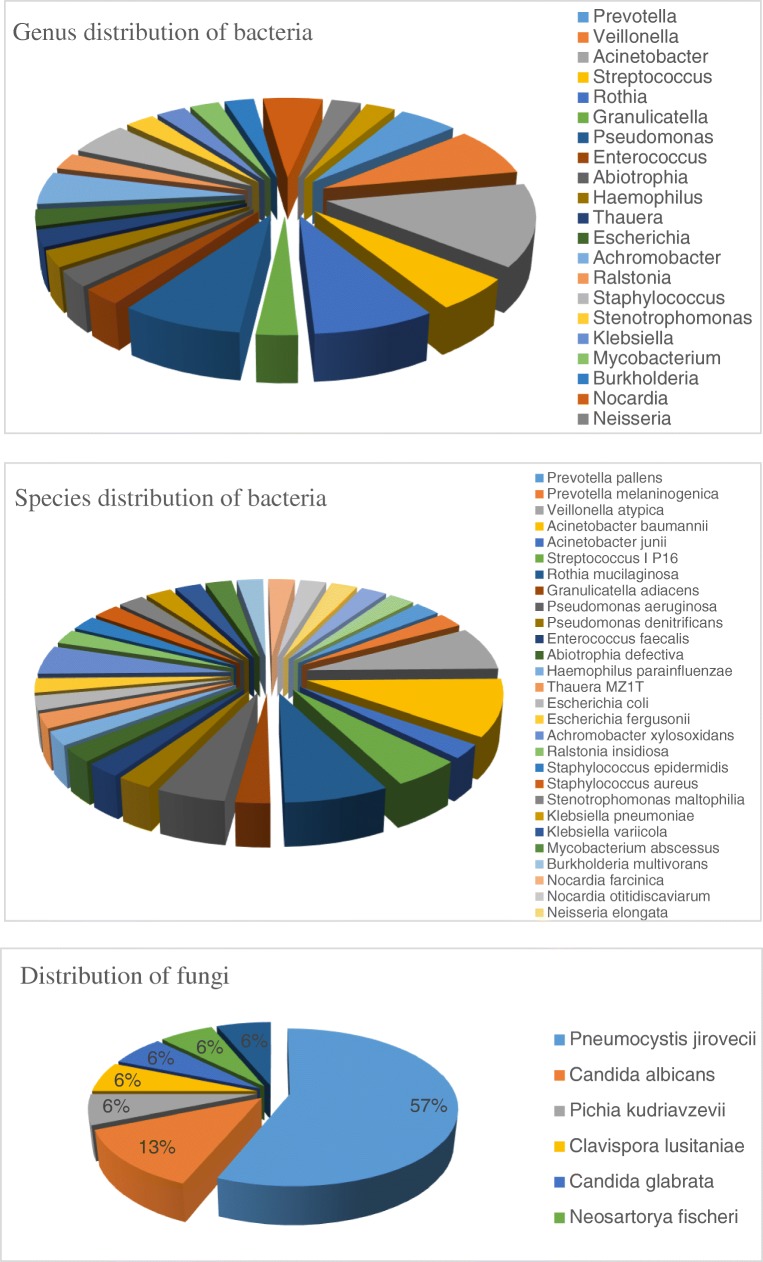
Figure 1 lists the distribution of bacteria and fungi identified by mNGS, among which Acinetobacter, Veillonella, Pseudomonas, and Rothia species were the most common bacteria. The most common fungus was Pneumocystis jirovecii (57%).
Fig. 1.
Genus and species distribution of bacteria and distribution of fungi by mNGS for the constituents for which the number of unique reads was ≥ 50
Figure 1 only lists the distribution of pathogens that met the definition of infection. The mNGS analysis identified the distribution of a number of bacteria with < 50 reads, which may indirectly reflect diversity of bacterial and fungal flora in the lower respiratory tract. Linear regression analysis showed that the diversity was significantly correlated with the interval between disease onset and sampling but not with underlying lung disease, smoking history, or interval from tracheal intubation to sampling (Fig. 2).
Fig. 2.
Number of bacterial and fungal genera identified in the bronchoalveolar lavage sample relative to the number of days for which the disease onset occurred before collection of the sample
Discussion
BAL is well tolerated and easily and safely performed in acutely ill patients (e.g. patients with ARDS) [14]. mNGS is suitable for the detection of pathogens that cannot be identified by other existing detection techniques, also in cases where patients fail to respond to standardized antimicrobial treatment. For rare and slow-growing bacteria, mNGS offers considerable advantages in shortening the time required for a diagnosis confirmation of bacterial/fungal infection, promoting targeted antimicrobial treatment, and improving patient prognosis [15]. However, mNGS findings should be combined with epidemiological and clinical characteristics before a pathogenic microbe can be identified.
The application of mNGS remains controversial due to technical aspects (e.g. methods to eliminate the impacts of host genes) and the interpretation of results. Thus far, most articles on mNGS focus on its diagnostic value for specific bacterial infections [12, 16]; minimal literature is available regarding accurate interpretation of all detected results. Based on the literature [3, 6], our current study analysed DNA levels of microbes and their levels of relative abundance in clinical BALF samples to identify pathogenic bacteria. This method significantly improved the sensitivity of pathogenic detection and provided guidance in clinical practice.
According to our results, positive rates of mNGS for identifying Mycobacterium tuberculosis and non-tuberculous mycobacteria were low due to their high homology [3]. Xpert MTB/RIF showed a positive result in one patient and another patient for tuberculosis on acid-fast staining of a sputum smear, suggesting the presence of non-tuberculous mycobacteria; however, the results were negative by mNGS.
The treatment strategies were changed in only 11 of the 32 patients. The limited value of mNGS may be because standardized anti-PC and anti-CMV treatments are typically applied immediately upon admission in our department. Furthermore, most patients admitted to the RICU have severe pneumonia, which is treated empirically for a wide range of pathogens. However, traditional methodologies combined with mNGS can better guide the discontinuation of antimicrobial therapy. High agreement rates between conventional methods and mNGS in our current study further verified the validity and accuracy of mNGS. For hospitals with limited detection capabilities, mNGS is recommended based on local epidemiological characteristics, especially for immunocompromised patients and those with severe or complicated infections.
Based on the number of reads and relative species abundance, the mNGS technique can be used for semi-quantitative detection. Two of our patients (no. 8 and no. 14) underwent BALF mNGS twice, and dynamic changes in the PC and adenovirus reads could indirectly reflect therapeutic effectiveness, which is a potential added value of this technique [17, 18]. Furthermore, mNGS can be used for adenovirus genotyping, thereby providing better clinical practice guidance. However, due to difficulties in virus culture and high false-positive rates of nucleic acid detection alone, few reports have described whether the detected DNA virus is a pathogenic microbe [18, 19]. The reads and rates of relative abundance determined by NGS may provide a quantitative basis for identification of viral infection; however, additional experiments are needed to verify the optimal cut-off values.
NGS can also identify the distribution of airway flora under different conditions. The literature indicates that airway flora composition is not related to smoking history [20] but is affected by duration of mechanical ventilation [21]. Our current study showed an association between interval from disease onset to sampling and species within the airway flora. The lower respiratory tract is a relatively sterile anatomical location; with prolonged onset time, the types of bacterial flora may change, which may be a result of bacterial flora migration. Loss of diversity within respiratory tract microbial communities is proposed as an ecological marker of infection [22, 23].
Our study was limited by a small sample size and diverse diseases in the patients sampled. Furthermore, neither genome-wide RNA sequencing nor drug resistance testing was performed. Although NGS has been widely accepted and applied in critically ill patients, there is no uniform standard to modify or guide clinical treatment strategies, especially for some intractable cases, which limits its application in clinical research.
In future studies, we will apply mNGS for the detection of inflammatory mediators or immune-related factors and drug resistance as well as for the detection and identification of aetiological agents.
Electronic supplementary material
(PDF 301 kb)
(PDF 761 kb)
Acknowledgements
The authors wish to thank the patients for participating in this study.
Author contributions
Conceptualization, Zhao-hui Tong; data curation, Ying Li; formal analysis, Ya-lan Liu; project administration, Bing Sun, Xiao Tang, Hang-yong He, and Xu-yan Li; resources, Rui Wang; software, Fei Guo; supervision, Zhao-hui Tong; writing (original draft), Ying Li; writing (review and editing), Bing Sun and Zhao-hui Tong
Funding information
This work was supported by the Capital’s Funds for Health Improvement and Research (2016–1-1061).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ying Li and Bing Sun contributed equally to this work.
References
- 1.Guo Ling-Yun, Feng Wen-Ya, Guo Xin, Liu Bing, Liu Gang, Dong Jie. The advantages of next-generation sequencing technology in the detection of different sources of abscess. Journal of Infection. 2019;78(1):75–86. doi: 10.1016/j.jinf.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 2.Liu P, Weng X, Zhou J, Xu X, He F, Du Y, et al. Next generation sequencing based pathogen analysis in a patient with neurocysticercosis: a case report. BMC Infect Dis. 2018;18:113. doi: 10.1186/s12879-018-3015-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li H, Gao H, Meng H, Wang Q, Li S, Chen H, et al. Detection of pulmonary infectious pathogens from lung biopsy tissues by metagenomic next-generation sequencing. Front Cell Infect Microbiol. 2018;8:205. doi: 10.3389/fcimb.2018.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leo Stefano, Gaïa Nadia, Ruppé Etienne, Emonet Stephane, Girard Myriam, Lazarevic Vladimir, Schrenzel Jacques. Detection of Bacterial Pathogens from Broncho-Alveolar Lavage by Next-Generation Sequencing. International Journal of Molecular Sciences. 2017;18(9):2011. doi: 10.3390/ijms18092011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer KC, Raghu G, Baughman RP, Brown KK, Costabel U, du Bois RM, et al. An official American Thoracic Society clinical practice guideline: the clinical utility of bronchoalveolar lavage cellular analysis in interstitial lung disease. Am J Respir Crit Care Med. 2012;185:1004–1014. doi: 10.1164/rccm.201202-0320ST. [DOI] [PubMed] [Google Scholar]
- 6.Langelier C, Zinter MS, Kalantar K, Yanik GA, Christenson S, O’Donovan B, et al. Metagenomic sequencing detects respiratory pathogens in hematopoietic cellular transplant patients. Am J Respir Crit Care Med. 2018;197:524–528. doi: 10.1164/rccm.201706-1097LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Force, ARDS Definition Task. Ranieri VM, Rubenfeld GD, Thompson BT, Ferguson ND, et al. Acute respiratory distress syndrome: the Berlin Definition. JAMA. 2012;307:2526–2533. doi: 10.1001/jama.2012.5669. [DOI] [PubMed] [Google Scholar]
- 8.Azoulay E, Pickkers P, Soares M, Perner A, Rello J, Bauer PR, et al. Acute hypoxemic respiratory failure in immunocompromised patients: the Efraim multinational prospective cohort study. Intensive Care Med. 2017;43(12):1808–1819. doi: 10.1007/s00134-017-4947-1. [DOI] [PubMed] [Google Scholar]
- 9.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-32.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cabello H, Torres A, Celis R, El-Ebiary M, Puig de la Bellacasa J, Xaubet A, et al. Bacterial colonization of distal airways in healthy subjects and chronic lung disease: a bronchoscopic study. Eur Respir J. 1997;10:1137–1144. doi: 10.1183/09031936.97.10051137. [DOI] [PubMed] [Google Scholar]
- 11.Byrd AL, Belkaid Y, Segre JA. The human skin microbiome. Nat Rev Microbiol. 2018;16:143–155. doi: 10.1038/nrmicro.2017.157. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Yi, Ai Jing-Wen, Cui Peng, Zhang Wen-Hong, Wu Hong-Long, Ye Ming-Zhi. A cluster of cases of pneumocystis pneumonia identified by shotgun metagenomics approach. Journal of Infection. 2019;78(2):158–169. doi: 10.1016/j.jinf.2018.08.013. [DOI] [PubMed] [Google Scholar]
- 13.Zinter Matt S, Dvorak Christopher C, Mayday Madeline Y, Iwanaga Kensho, Ly Ngoc P, McGarry Meghan E, Church Gwynne D, Faricy Lauren E, Rowan Courtney M, Hume Janet R, Steiner Marie E, Crawford Emily D, Langelier Charles, Kalantar Katrina, Chow Eric D, Miller Steve, Shimano Kristen, Melton Alexis, Yanik Gregory A, Sapru Anil, DeRisi Joseph L. Pulmonary Metagenomic Sequencing Suggests Missed Infections in Immunocompromised Children. Clinical Infectious Diseases. 2018;68(11):1847–1855. doi: 10.1093/cid/ciy802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hertz MI, Woodward ME, Gross CR, Swart M, Marcy TW, et al. Safety of bronchoalveolar lavage in the critically ill, mechanically ventilated patient. Crit Care Med. 1991;19:1526–1532. doi: 10.1097/00003246-199112000-00015. [DOI] [PubMed] [Google Scholar]
- 15.Seo S, Renaud C, Kuypers JM, Chiu CY, Huang ML, Samayoa E, et al. Idiopathic pneumonia syndrome after hematopoietic cell transplantation: evidence of occult infectious etiologies. Blood. 2015;125:3789–3797. doi: 10.1182/blood-2014-12-617035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ai JW, Weng SS, Cheng Q, Cui P, Li YJ, Wu HL, et al. Human endophthalmitis caused by pseudorabies virus infection, China, 2017. Emerg Infect Dis. 2018;24:1087–1090. doi: 10.3201/eid2406.171612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ai JW, Zhang HC, Cui P, Xu B, Gao Y, Cheng Q, et al. Dynamic and direct pathogen load surveillance to monitor disease progression and therapeutic efficacy in central nervous system infection using a novel semi-quantitive sequencing platform. J Infect. 2018;76:307–310. doi: 10.1016/j.jinf.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 18.Hu Y, Zhang Y, Ren X, Liu Y, Xiao Y, Li L, et al. A case report demonstrating the utility of next generation sequencing in analyzing serial samples from the lung following an infection with influenza A (H7N9) virus. J Clin Virol. 2016;76:45–50. doi: 10.1016/j.jcv.2015.12.013. [DOI] [PubMed] [Google Scholar]
- 19.Petty TJ, Cordey S, Padioleau I, Docquier M, Turin L, Preynat-Seauve O, et al. Comprehensive human virus screening using high-throughput sequencing with a user-friendly representation of bioinformatics analysis: a pilot study. J Clin Microbiol. 2014;52:3351–3361. doi: 10.1128/JCM.01389-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morris A, Beck JM, Schloss PD, Campbell TB, Crothers K, Curtis JL, et al. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am J Respir Crit Care Med. 2013;187:1067–1075. doi: 10.1164/rccm.201210-1913OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huebinger RM, Liu MM, Dowd SE, Rivera-Chavez FA, Boynton J, Carey C, et al. Examination with next-generation sequencing technology of the bacterial microbiota in bronchoalveolar lavage samples after traumatic injury. Surg Infect (Larchmt) 2013;14:275–282. doi: 10.1089/sur.2012.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahashi Y, Saito A, Chiba H, Kuronuma K, Ikeda K, Kobayashi T, et al. Impaired diversity of the lung microbiome predicts progression of idiopathic pulmonary fibrosis. Respir Res. 2018;19:34. doi: 10.1186/s12931-018-0736-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abreu NA, Nagalingam NA, Song Y, Roediger FC, Pletcher SD, Goldberg AN, et al. Sinus microbiome diversity depletion and Corynebacterium tuberculostearicum enrichment mediates rhinosinusitis. Sci Transl Med. 2012;4:151ra124. doi: 10.1126/scitranslmed.3003783. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 301 kb)
(PDF 761 kb)