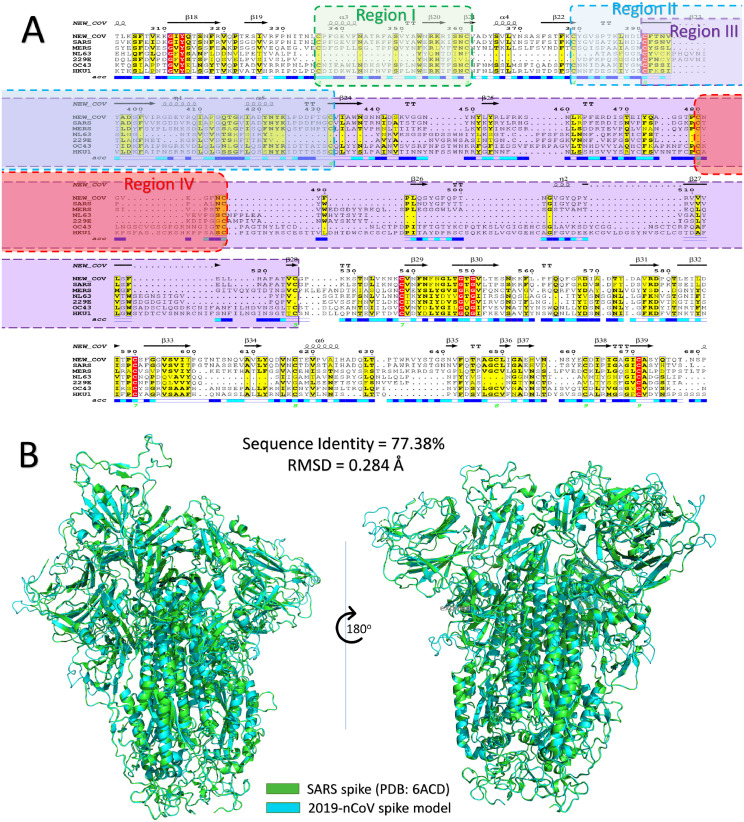
Fig. 1.
(A) Part of the multiple sequence alignment for the spike protein of all of the currently reported human coronaviruses strains (COVID-19, SARS, MERS, NL63, 229E, OC43, and HKU1). The alignment is made using the Clustal Omega web server and is displayed by ESpript 3 software. The red highlighted residues are identical, while yellow highlighted residues are conserved among the seven HCoVs. Secondary structures are represented at the top of the MSA for the COVID-19 spike, while the surface accessibility is shown at the bottom (blue, surface accessible, cyan, partially accessible, and white for buried residues). The four regions of the spike protein are shaded with green, blue, magenta, and red for regions I, II, III, and IV, respectively. (B) Structural superposition of SARS spike structure (green cartoon) and COVID-19 spike model (cyan cartoon). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)