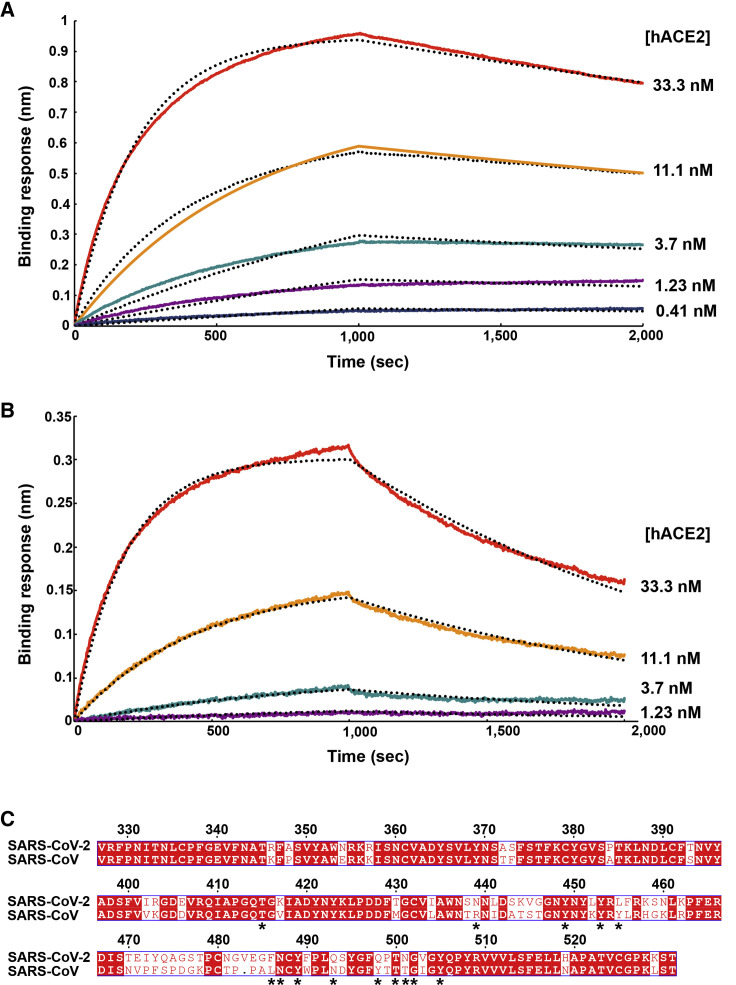
Figure 2.
SARS-CoV-2 S Recognizes hACE2 with Comparable Affinity to SARS-CoV S
(A and B) Biolayer interferometry binding analysis of the hACE2 ectodomain to immobilized SARS-CoV-2 SB (A) or SARS-CoV SB (B). The experiments were repeated with different protein preparations and one representative set of curves is shown. Dotted lines correspond to a global fit of the data using a 1:1 binding model.
(C) Sequence alignment of SARS-CoV-2 SB and SARS-CoV SB Urbani (late phase of the 2002–2003 SARS-CoV epidemic). Identical and similar positions are respectively shown with white or red font. The single amino acid insertion at position 483 of the SARS-CoV-2 SB is indicated with a period at the corresponding SARS-CoV SB position. The 14 residues that are key for binding of SARS-CoV SB to hACE2 are labeled with a star. See also Data S1.