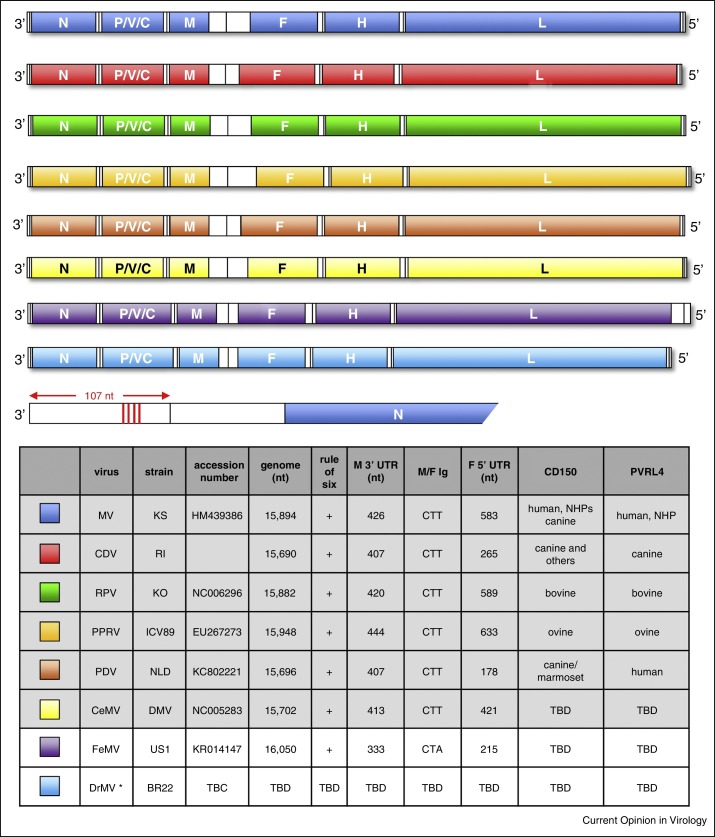
Figure 1.
Schematic representation of the genomic organization of known and proposed morbilliviruses. MV = measles virus (blue), CDV = canine distemper virus (red), RPV = rinderpest virus (green), PPRV = peste des petits ruminants virus (light orange), PDV = phocine distemper (dark orange), CeMV = cetacean morbilliviruses (yellow), FeMV = feline morbillivirus (purple) and DrMV = vampire bat morbillivirus (light blue). Genomes are drawn to relative scale for comparison of the 3′ and 5′ termini, open reading frames and intergenic sequence length. Open reading frames represent the nucleocapsid (N), phospho- (P), matrix (M), fusion (F), hemagglutinin (H) and large (L) proteins which are present in the virions. The P/V/C gene encodes the non-structural V and C proteins. Four conserved hexamers in the 107 nucleotide (nt) 3′ leader sequence of MV are indicated (vertical red lines). The table lists known (gray) and proposed (white) morbilliviruses. Standard genome lengths, representative strains, proven CD150 and PVRL4 receptor use, 3′ M gene untranslated region (UTR) lengths, the sequence of the non-transcribed intergenic (Ig) trinucleotide spacer between the M and F genes, the length of the 5′ UTR of the F gene and available accession numbers on which the schematic diagrams, genome lengths etc. are based. Colors from this plate are used equivalently in figure 2 for clarity and comparability. * DrMV has not been isolated as a replicating virus.