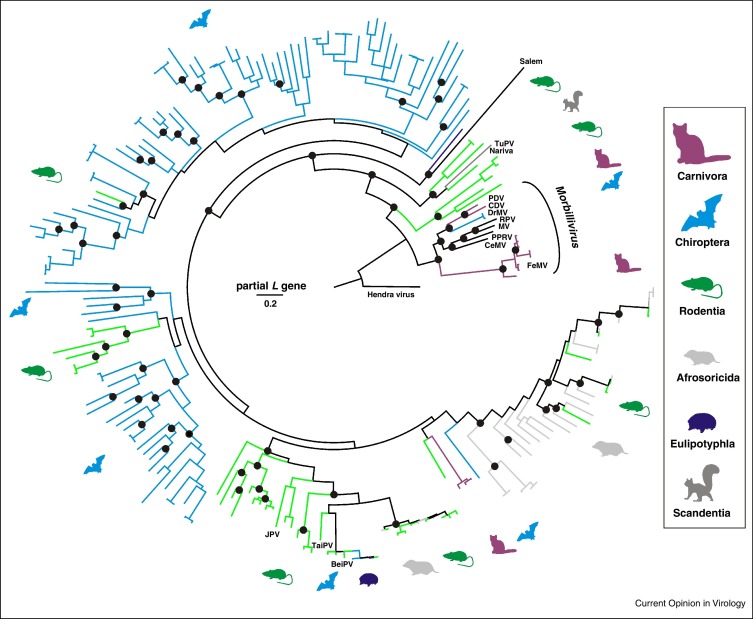
Figure 3.
Phylogenetic relationships of morbilliviruses and related small mammal viruses. Bayesian phylogenetic reconstruction done using MrBayes V3.1 with 2 000 000 tree iterations sampled every 100 steps, corresponding to 20 000 trees of which 25% were discarded as burn-in. The final tree was annotated using TreeAnnotator and visualized using FigTree from the BEAST package. Hendra virus was used as an outgroup. Host orders are indicated by color and pictograms to the right. Prototype morbilliviruses and related viruses are indicated next to branches (JPV, J virus; BeiPV, Beilong virus; TuPV, Tupaia paramyxovirus; see text for other abbreviations). All available unclassified morbilli-related viruses (UMRV) that differed by more than 5% in their partial L gene sequences from any other virus, were from different hosts or different sampling countries were included into the analysis. The final dataset comprised 205 partial L gene sequences of 438 nucleotides generated by the RT-PCR assay described by [93] commonly used in paramyxovirus field studies.