Figure 1.
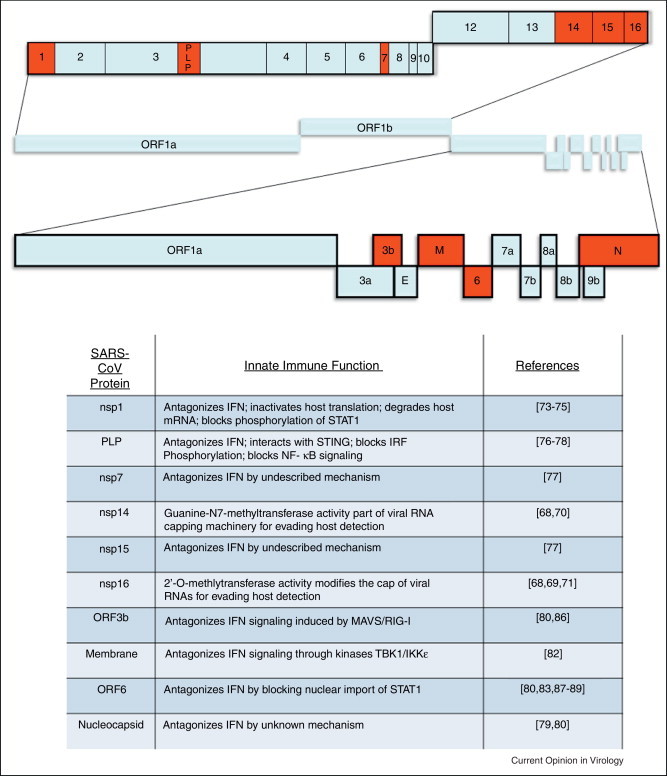
The SARS-CoV genome and functions of SARS-CoV innate immune antagonists. (a) The typical coronavirus genome size is quite large in comparison to many other positive-sense RNA viruses; within the SARS-CoV genome of 29.7 kB at least ten genes with potential functions that modulate innate immunity have been characterized (highlighted here in red). Like other members of the viral family Coronaviridae, SARS-CoV has a positive-sense, single-stranded RNA genome that is amenable to manipulation using reverse genetic techniques [90]. In SARS-CoV the first open reading frame (ORF) encodes the 16 nonstructural proteins that make up the viral replicase, while the ensuing ORFs encode four structural proteins that compose the virion, as well as eight accessory proteins. The SARS-CoV accessory proteins share no homology to the accessory proteins of other human coronaviruses, and while dispensable for replication in vitro, encode functions that probably impact viral pathogenesis in vivo [91]. While SARS-CoV was a novel virus not previously recognized before the 2002 outbreak, other coronaviruses have been associated with disease in humans. Coronaviruses known to infect humans include HCoV-HKU1, HCoV-OC43, HCoV-229E, and HCoV-NL63, which also cause respiratory infections but are generally much less severe than SARS [92]. (b) Transcription and subsequent signaling of Interferon is vital for activating the antiviral response in host cells. Because of this, many viruses (including SARS-CoV) encode proteins that antagonize the IFN response to viral infection. Of the SARS-CoV viral proteins listed here, eight have been identified as Interferon antagonists and two have been implicated in the viral RNA capping machinery.
