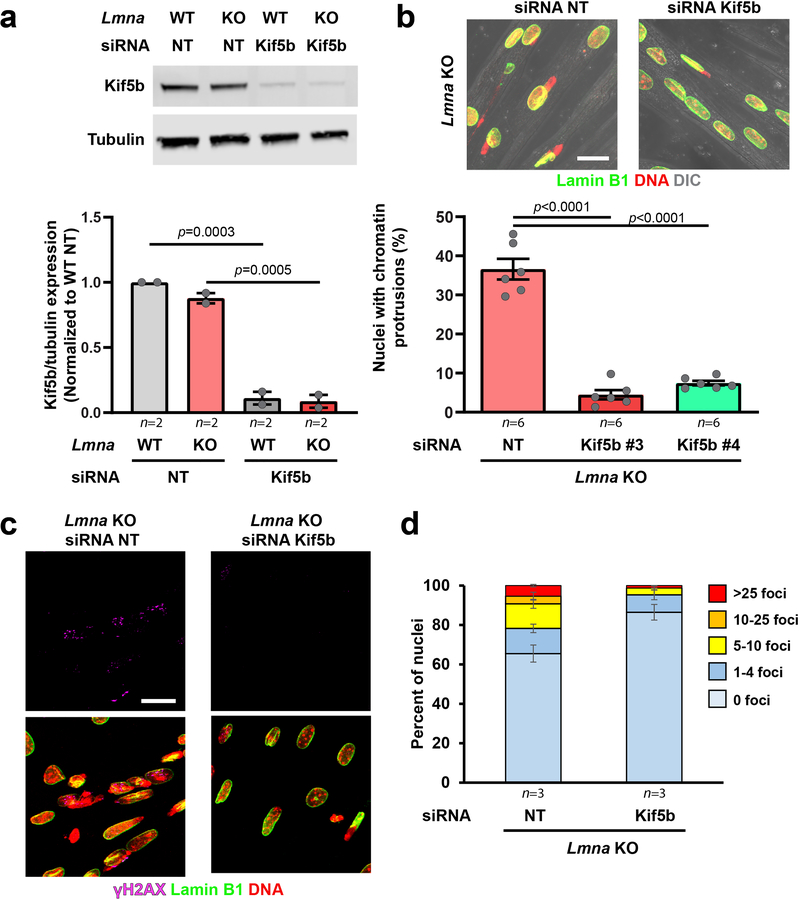
Extended Data Fig. 8. Kif5b depletion in myotubes reduced chromatin protrusions and DNA damage in Lmna KO myonuclei.
(a) Western blot for Kif5b in myoblasts treated with a non-target control siRNA (siRNA NT) or siRNA against Kif5b. Data based on n independent experiments. (Bottom) Corresponding quantification. Significance determined by two-tailed students t-test within each genotype. (b) Representative images of Lmna KO myofibers at day 5 of differentiation treated with either a non-target control siRNA (siRNA NT) or siRNA against kinesin-1 (siRNA Kif5b) at day 0. Scale bar: 20 μm. Quantification of the number of chromatin protrusions at day 5 of differentiation in Lmna KO cells treated with non-target (NT) siRNA or depleted for Kif5b using two independent siRNAs (Kif5b #3 and Kif5b #4). Data based on n independent experiments, with 155–270 nuclei counted per experiment. Significance determined by one-way ANOVA, using Tukey’s correction for multiple comparisons. (c) Representative images of Lmna KO cells treated with either non-target (NT) siRNA or siRNA against Kif5b and immunofluorescently labeled for γH2AX, showing fewer chromatin protrusions and less DNA damage in the Kif5b depleted cells. Experiments were conducted three independent times, with similar results. Scale bar: 20 μm. (d) Quantification of the number of γH2AX foci in Lmna KO myonuclei following treatment with either non-target siRNA or siRNA against Kif5b. Data based on n independent experiments, in which 27–53 nuclei were counted per experiment. All bar plots show mean value ± standard error of the mean.