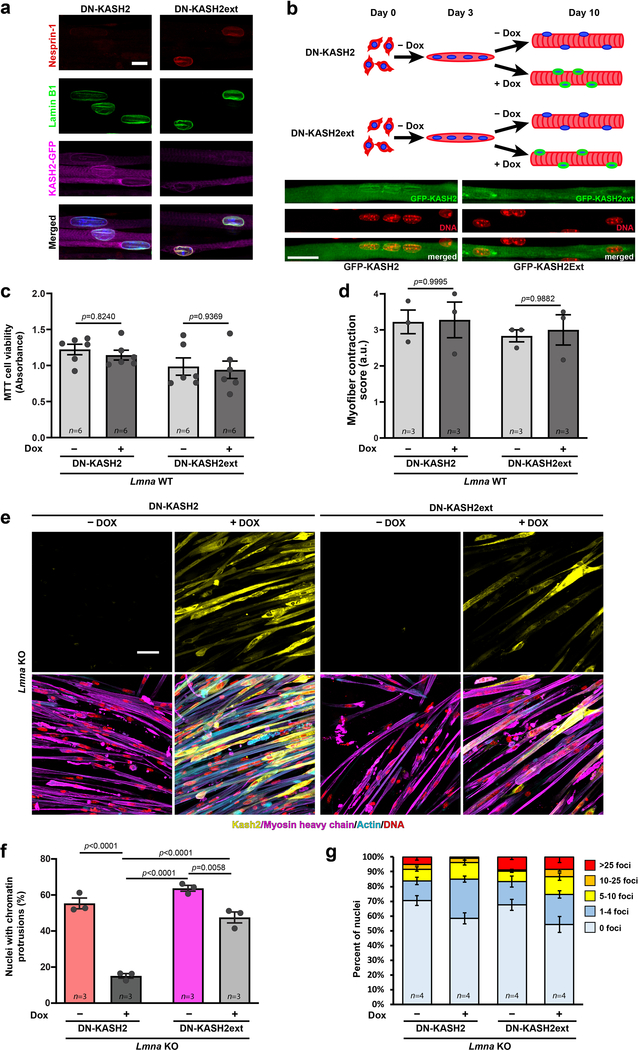
Extended Data Fig. 9. Expression of the DN-KASH2 construct disrupts the LINC complex and limits nuclear movement, without affecting myofiber function in Lmna WT myofibers.
(a) Representative image showing displacement of endogenous nesprin-1 in myofibers expressing the DN-KASH2 construct, and no displacement of nesprin-1 in myofibers expressing the DN-KASH2ext construct. Scale bar: 10 μm. (b) Representative image showing nuclear clustering in myofibers expressing the DN-KASH2 construct, and normal nuclear spreading in myofibers expressing the DN-KASH2ext construct. Scale bar: 20 μm. (c) Quantification of cell viability following DN-KASH2 or DN-KASH2ext treatment in Lmna WT cells using the MTT assay. Data based on n differentiation replicates per condition, from three independent experiments. Significance determined by two-way ANOVA (genotype; DOX treatment), using Tukey’s correction for multiple comparisons. (d) Quantification of myofiber contraction following DN-KASH2 or DN-KASH2ext treatment in Lmna WT cells based on the percent of contractile fibers. Data based on n independent experiments, for which scores were determined based on 5–6 image sequences per condition. Significance determined by two-way ANOVA (genotype x DOX treatment), using Tukey’s correction for multiple comparisons. (e) Representative images of Lmna KO expressing either DN-KASH2 or DN-KASH2ext, with or without 1 μM doxycycline (DOX), and immunofluorescently labeled for myosin heavy chain, actin, and DNA (DAPI) showing increased cell area and enhanced sarcomeric staining in the DOX treated cells expressing DN-KASH2. Scale bar: 50 μm. (f) Quantification of the number of chromatin protrusions in Lmna KO myonuclei expressing either DN-KASH2 or DN-KASH2ext. Data based on n independent experiments per condition. Significance determined by two-way ANOVA (genotype; DOX treatment), using Tukey’s correction for multiple comparisons. (g) Quantification of the extent of DNA damage based on the number of γH2AX foci per nucleus during myofiber differentiation. Lmna KO myonuclei expressing the DN-KASH2 construct show a decrease in the nuclei with severe DNA damage (>25 foci). Data based on n independent experiments per condition. All bar plots show mean value ± standard error of the mean.