Figure 5.
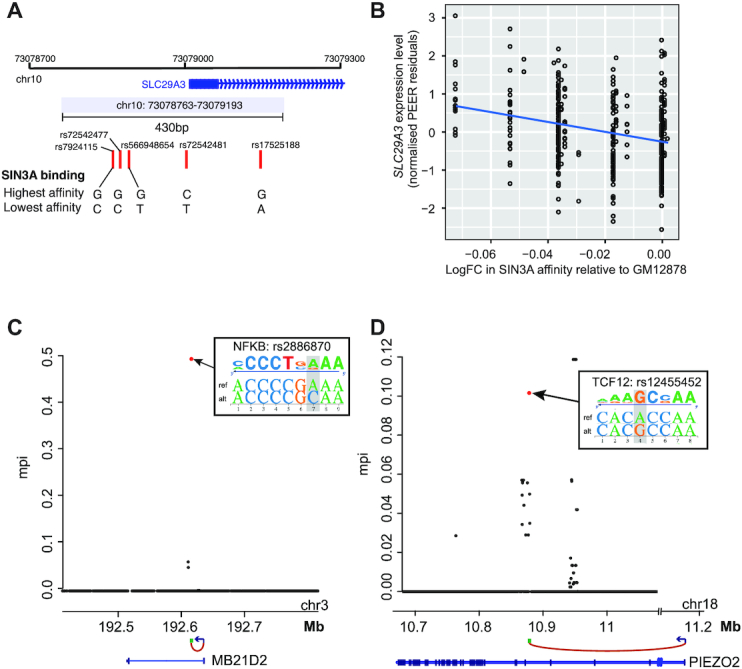
Unthresholded approach for detecting TF binding affinity CRM variant associations with gene expression and their validation using GUESSFM. (A) Example of a CRM with multiple SNPs affecting the affinity for the same TF, SIN3A. (B) Association between log-fold change in CRM affinity for SIN3A (relative to the highest affinity allele observed in GM12878) and mRNA level (normalized PEER residuals) of the connected gene, SLC29A3 (gene-level FDR adjusted P-value = 1.60 × 10−4, β = −0.14). (C, D) Examples of loci, whereby the SNP predicted to have the strongest impact on a CRM’s binding affinity for a given TF has been fine-mapped as a potentially causal variant driving the locus’s association with the expression of a physically connected target gene (GUESSFM  ). (C) eGene: MB21D2; eQTL rs2886870, predicted to affect NFKB binding affinity. (D) eGene: PIEZO2; eQTL rs12455452, predicted to affect TCF12 binding affinity. See insets for the effects of the SNPs on the respective TF's PWM match.
). (C) eGene: MB21D2; eQTL rs2886870, predicted to affect NFKB binding affinity. (D) eGene: PIEZO2; eQTL rs12455452, predicted to affect TCF12 binding affinity. See insets for the effects of the SNPs on the respective TF's PWM match.
