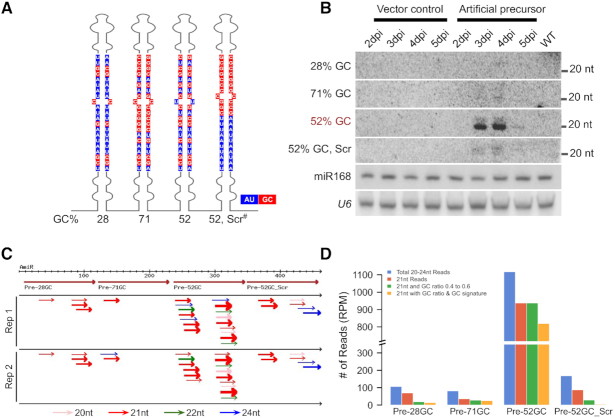
Figure 3.
Efficient GC-content-specific processing of miRNA precursors in vivo. (A) 2D representation of artificial precursor with four stem-loops, each having unique mature sequences of varying GC content as indicated. The highlighted region represents miRNA/miRNA* region. AU marked in blue and GC in red. This construct was expressed under the CaMV 35S promoter. ‘Scr#’ - scrambled signature. (B) Northern analysis of miRNAs with different GC content in N. tabacum leaves. U6 and miR168 served as loading controls. (C) Sequenced sRNAs from N. tabacum leaves expressing artificial precursor at 4 dpi, were mapped to the precursor. Four big brown arrows indicate the four stem-loops in tandem. Pink, red, green and blue represent 20, 21, 22 and 24-nt reads, respectively. The thickness of the arrow is indicative of the abundance. Only reads sequenced more than 5 RPM are considered. (D) Graph comparing the abundance of sRNAs mapped to the artificial precursor. Blue (all reads of 21–24-nt), red (21-nt), green (reads that have GC content between 40% and 60%), yellow (reads that have GC content between 40% and 60% and also GC signature) bars indicate abundance.