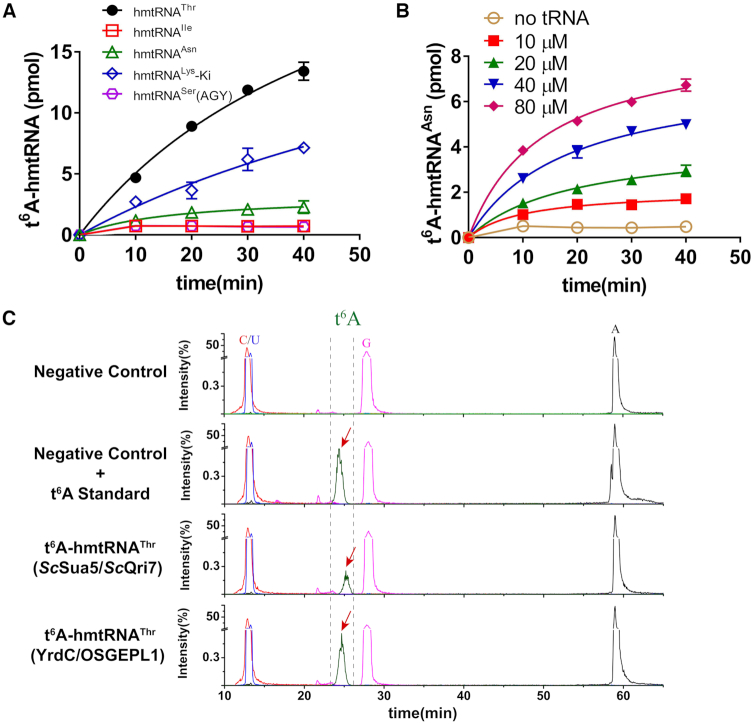
Figure 3.
The hmtRNAThr transcript is the best substrate of YrdC/OSGEPL1 in vitro. (A) Time-course curves of the modification of five hmtRNAs, hmtRNAAsn (green triangles), hmtRNAIle (red squares), hmtRNALys (blue diamonds), hmtRNASer(AGY) (pink hexagons) and hmtRNAThr (black filled circles), by YrdC and OSGEPL1. (B) t6A modification at increasing concentrations of hmtRNAAsn (10 μM, red filled squares; 20 μM, green filled triangles; 40 μM, blue filled inverted triangles and 80 μM, pink filled diamonds). A control without tRNA addition (no tRNA, orange circles) was included. (C) LC–MS/MS analysis of the digestion product of the hmtRNAThr transcript in the absence and presence of standard t6A or modified hmtRNAThr transcripts by YrdC/OSGEPL1 or Sua5/Qri7.