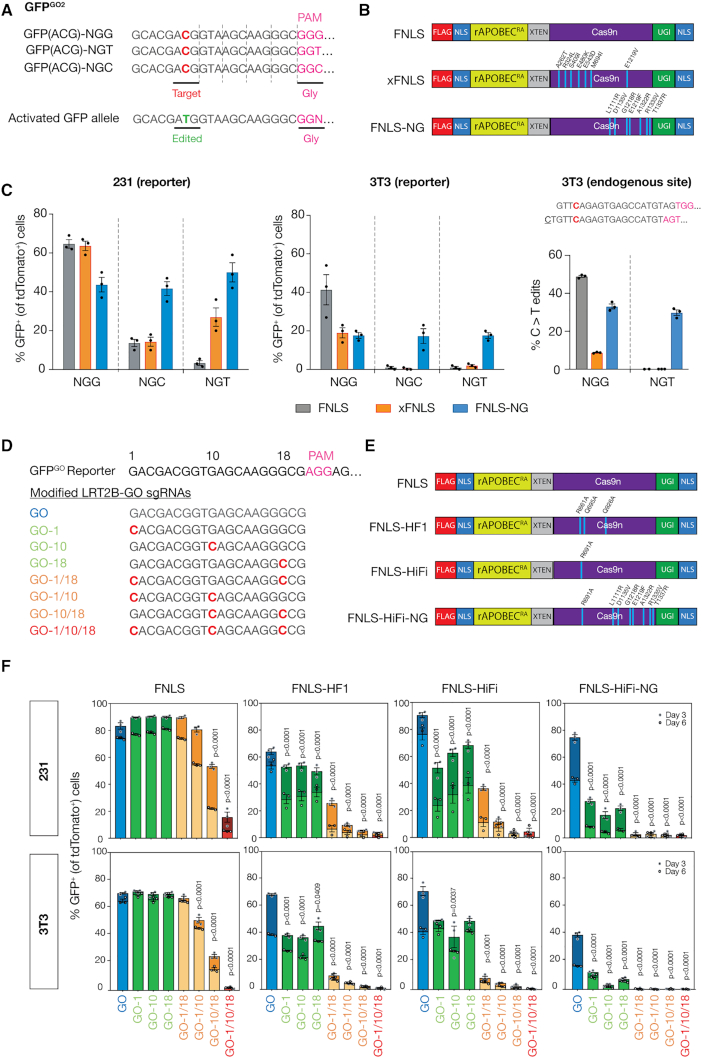
Figure 2.
GO quantifies base editing activities of BE enzymes with various PAM specificities and fidelities. (A) Panel of GFPGO2 reporters with different PAM recognition sequences: NGG, NGT, NGC designed with constant 20 bp complementary target sequence at 5′ end of GFP cDNA for targeting by sgGO2-tdTomato. (B) Schematic of BE PAM variant enzymes included in experimental panel. (C) Flow cytometry analysis of GFP activation in (left) 231 and (middle) 3T3 cell lines expressing the panel of editors (B) and infected with each of the PAM GFPGO2 reporters 6 days after transduction with sgGO2 (notated by GFPGO2 PAM site). GFP+ cells were gated on tdTomato+ population. (right) Schematic showing the endogenous Apc locus (codon 1405) in 3T3 cells that contains a cytosine targetable by two nearly identical sgRNA with adjacent and distinct PAM recognition sequences, NGG and NGT. 3T3 cell lines expressing the panel of editors (B) were transduced with either of two nearly identical sgRNAs (NGG or NGT). Deep sequencing analysis of C>T editing events as a fraction of total reads at the Apc.1405 locus in the targeted panel of cells (bottom). (D) A panel of sgRNAs targeting the 5′ sequence of the GFPGO cDNA modified from sgGO were designed to include one to three mismatches across its complementary sequence. (E) Schematic of high fidelity BE enzymes included in experimental panel, including novel HiFi-NG enzyme created by combining mutations indicated to improve fidelity, and/or PAM flexibility. (F) 231 and 3T3 cells stably expressing GFPGO and panel of high-fidelity BE enzymes 3 and 6 days post-transduction with the panel of mismatch sgGOs in the LRT2B vector. GFP+ cells were gated on tdTomato+ cells.