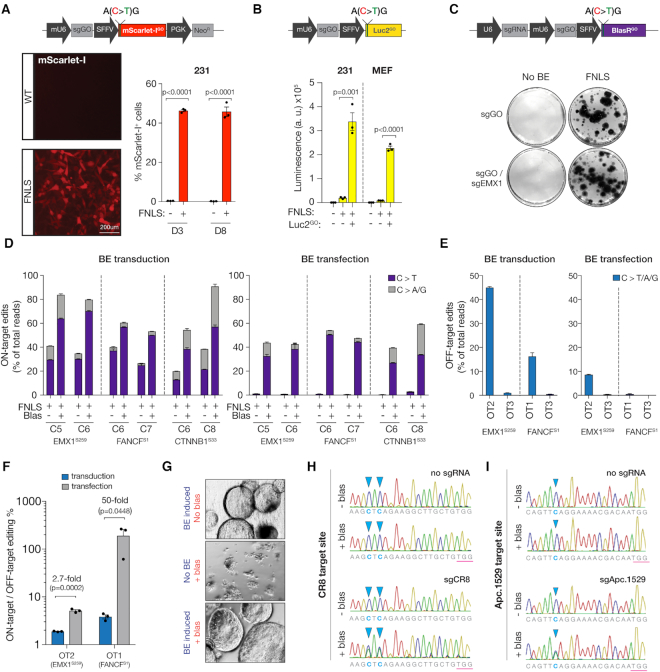
Figure 3.
GO is highly modular and can be functionally exploited to enrich base editing activity. (A) (top) Schematic of an all-in-one mScarlet-I GO reporter with (i) modified 5′ end in the mScarlet-I cDNA to include the silent start site ‘ACG’ within the 20bp sgGO complementary sequence, (ii) sgGO and (iii) neomycin selection gene (together, ScarGO). (bottom, left) Fluorescence microscopy of 231s expressing no editor (WT) or FNLS 3 days after infection with ScarGO. (bottom, right) Flow cytometry of 231s expressing no editor (WT) or FNLS 3- and 8-days after infection with ScarGO. (B) (top) Schematic of an all-in-one-Luciferase GO reporter with (i) a modified 5′ end in the Luciferase2 cDNA to include the silent start site ‘ACG’ within the 20 bp sgGO complementary sequence and (ii) sgGO (together, Luc2GO). (bottom) Luminescence in 231s and immortalized MEFs expressing no editor (WT) or FNLS 3 days after infection with and without Luc2GO. (C) (top) Schematic of an all-in-one Blasticidin GO reporter with (i) modified 5′ end in the Blasticidin (Blas) resistance gene to include silent start site ‘ACG’ within the 20-bp sgGO complementary sequence (ii) sgGO and (iii) an endogenous targeting sgRNA under an independent U6 promoter (together, BlasGO). (bottom) GEMSA staining/colony forming assay of Blas-treated DLD1 cells with FNLS or no editor (WT) infected with BlasGO containing sgRNAs targeting EMX1 or no sgRNA. (D) Deep sequencing analysis of corresponding loci quantifying editing events as a fraction of total reads in DLD1 cells with BlasGO (containing gRNAs EMX1.259, FANCF.S1, CTNNB1.S33) (left) transduced or (right) transfected with and without FNLS or Blas treatment. (E) Quantification of editing events for two off-target sites corresponding to two endogenous gRNAs EMX1.259, FANCF.S within BlasGO in DLD1 cells (left) transduced or (right) transfected with FNLS (F) Ratio of on target to off target editing events for the OT sites with editing >1% in DLD1 cells transduced or transfected with FNLS. (G) Brightfield images of murine pancreatic organoids with inducible base editor enzyme infected with BlasGO and treated with and without Blas. (H) Sanger sequencing chromatograms of CR8 locus in pancreatic organoids expressing BE enzyme and infected with BlasGO (containing no gRNA or sgRNA targeting CR8) with and without Blas treatment. (I) Sanger sequencing chromatograms of Apc.1529 locus in pancreatic organoids expressing BE enzyme and infected with BlasGO (containing no gRNA or sgRNA targeting Apc.1529) with and without Blas treatment.