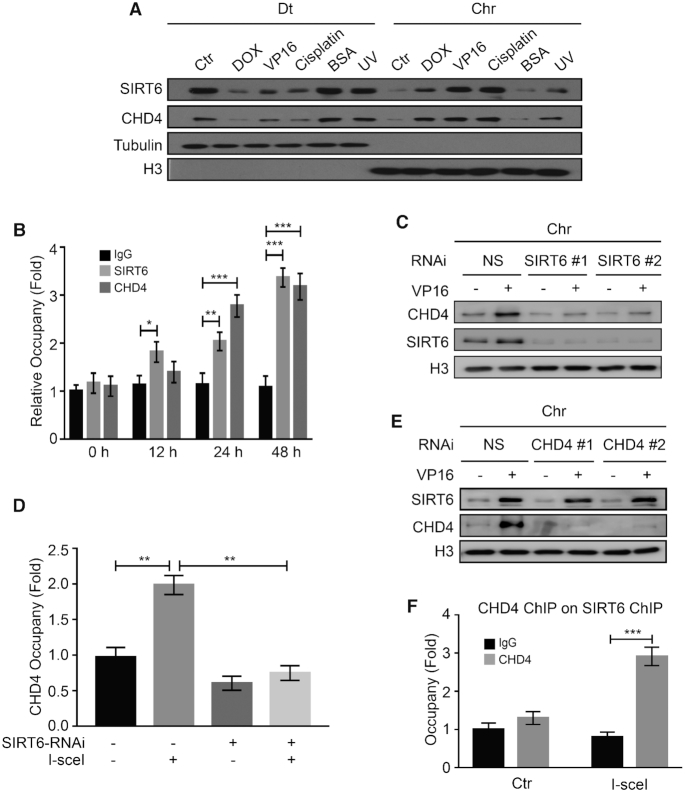
Figure 2.
SIRT6 is required to recruit CHD4 to chromatin following DNA damage. (A) HCT116 cells were treated with 1 μM DOX/40 μM VP16 /10 μM cisplatin/0.5% BSA for 0.5 h or treated with 200 J/m2 UV. The cells were fractionated (Dt/Chr) and analyzed by western blotting. (B) DR-U2OS cells were transfected with an I-SceI expression plasmid for up to 48 h. ChIP experiments were performed using an anti-SIRT6 or an anti-CHD4 antibody. SIRT6 and CHD4 binding to the DNA sequences flanking the I-SceI sites were measured. IgG was used as a negative control. Data are normalized to the control (no I-SceI) samples. (C) HCT116 cells were transfected with NS (non-specific) or two independent SIRT6 specific siRNAs for 48 h in the presence or absence of 40 μM VP16. Chromatin fractions were extracted and analyzed by western blotting. (D) DR-U2OS cells were transfected with NS or SIRT6-specific siRNA for 48 h; then, the cells were transfected with or without an I-SceI expression plasmid for 24 h. ChIP experiments were performed using an anti-CHD4 antibody. CHD4 binding to the DNA sequences flanking the I-SceI sites was measured. (E) HCT116 cells were transfected with NS or two independent CHD4-specific siRNAs for 48 h in the presence or absence of 40 μM VP16. Chromatin fractions were extracted and analyzed by western blotting. (F) DR-U2OS cells were transfected with or without the I-SceI expression plasmid for 48 h. Sequential ChIP-reChIP was performed on the I-SceI DNA damage sites by immunoprecipitation with an anti-SIRT6 antibody followed by immunoprecipitation with an anti-CHD4 antibody. The data represent the means ± SEM (n = 3, *P < 0.05, **P < 0.01, ***P < 0.001).