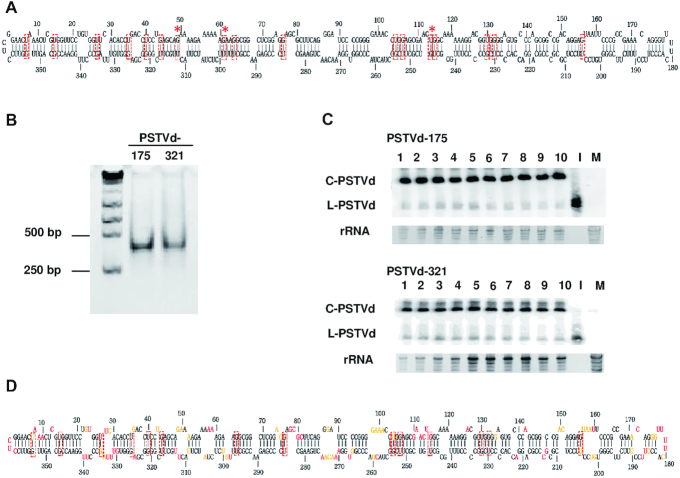
Figure 2.
Analysis of PSTVd G/U pairs by whole molecule SHAPE. (A) The canonical secondary structure of PSTVd containing 17 G/U pairs (red boxes) is shown. Red asterisks indicate G/U pairs not supported by whole molecule SHAPE (see 1D). (B) Folded, full-length PSTVd in vitro transcripts prepared for SHAPE, beginning at nucleotide positions 175 or 321, were tested for homogeneity by native polyacrylamide gel electrophoresis. A gel stained with ethidium bromide is shown. The positions of 250 bp and 500 bp size markers are indicated. (C) The infectivity of PSTVd-175 and PSTVd-321 transcripts was tested by inoculating 10 N. benthamiana plants with each RNA. Blots of RNA prepared from systemically infected leaves hybridized with a PSTVd-specific probe are shown. No signal was observed in RNA preparations from mock-inoculated plants (M). A full-length linear PSTVd in vitro transcript served as inoculum (I). Circular (C) and linear (L) forms of PSTVd RNAs are indicated. rRNA stained with ethidium bromide was a loading control. (D) The PSTVd secondary structure obtained by whole molecule SHAPE and folded with RNAstructure software is shown. Experiments were performed in triplicate with each RNA, and the averaged and similar data were used to derive the structure. The 14 G/U pairs predicted by this analysis are indicated by red boxes. SHAPE reactivity is indicated by nucleotide color: red = high reactivity (>0.85), orange = intermediate (0.40–0.85), black = low (0–0.40).