Figure 5.
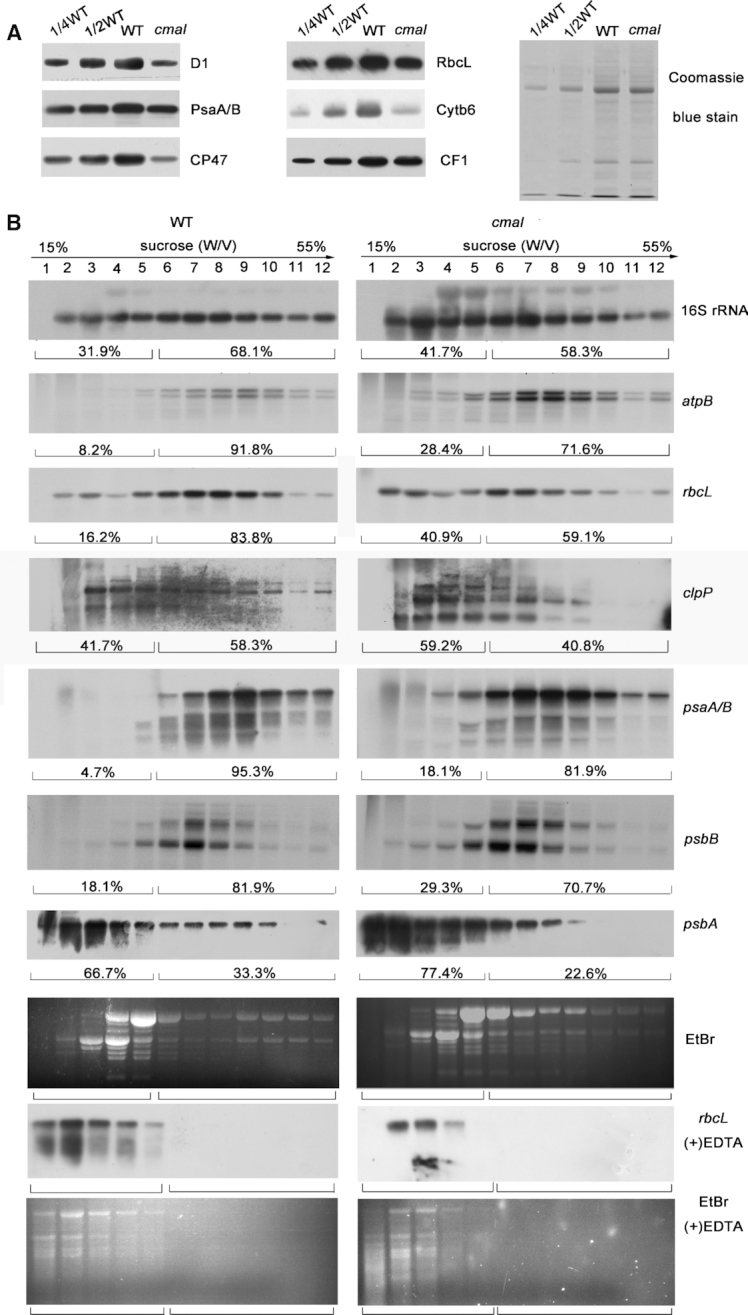
The translation efficiency of chloroplast mRNAs was reduced in cmal. (A) Immunoblot analysis of chloroplast-encoded proteins in cmal and WT plants. Total protein (10 μg) was extracted from four-week-old WT and cmal plants, separated using SDS-PAGE, and probed with different antibodies for specific proteins. The Coomassie blue-stained blot at the bottom serves as a loading control. (B) Polysome profiles of 4-week-old WT and cmal leaves. Twelve fractions of equal volume were collected from the top to bottom of 15% to 55% sucrose gradients. Equal proportions of the RNA purified from each fraction were analyzed using gel-blot hybridizations with the different probes. As a control, polysomes were isolated in the presence of EDTA which can disrupt polysome association, blotted, and hybridized to the rbcL probe. Signals of the polysomal fractions (fractions 1–5) and monosomes/free RNA fractions (fractions 6–12) were quantified with Image J respectively as described in Schult et al. (54). For the analysis of the rRNA distribution, an ethidium bromide-stained agarose gel prior to blotting is shown.
