Figure 3.
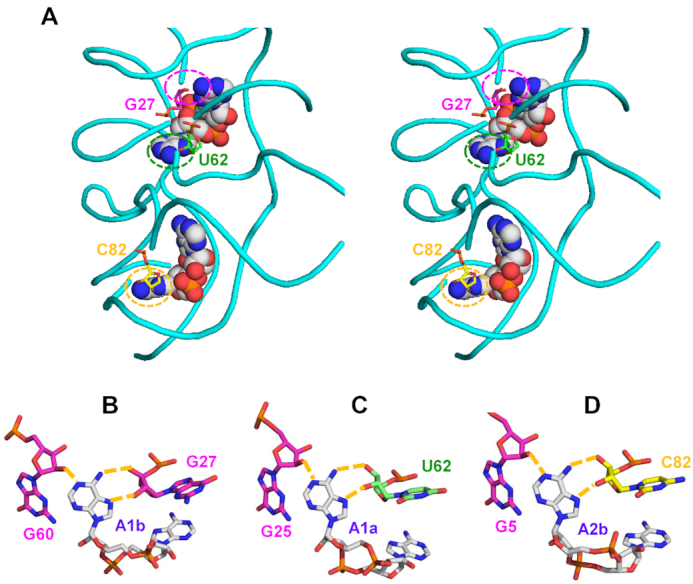
Binding of c-di-AMP to the YdaO riboswitch. (A) Stereo view of two c-di-AMP molecules (shown as spheres) bound to the YdaO riboswitch from B. subtilis (PDB: 4W90), whose sugar-phosphate backbone is shown in cyan (116). C-di-AMP-binding ribose moieties of G27, U62, and C82 are shown in stick representation and indicated by dashed circles. (B–D) Role of the ribose hydroxyl groups of the riboswitch in binding c-di-AMP (carbon atoms in grey). Hoogsteen side hydrogen bonds between N6 and N7 atoms of adenine bases of c-di-AMP and the ribose 2′- and 3′-hydroxyls from riboswitch bases G27 (B), U62 (C), and C82 (D) are coupled with adenine-N1 binding by 2′-hydroxyl groups of G60 (B), G25 (C) and G5(D). The abundance of hydrogen bonds explains the much higher affinity of c-di-AMP binding by the riboswitch than by protein receptors.
