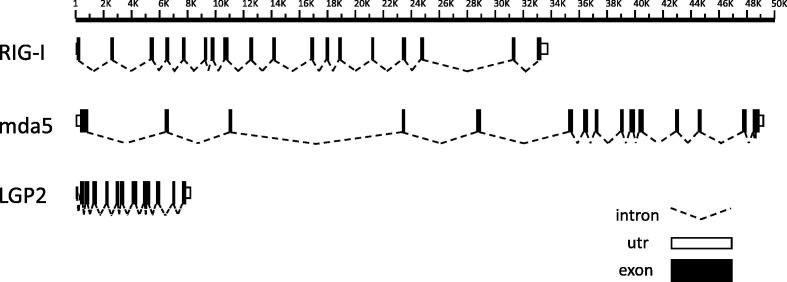
Fig. 1.
Predicted intron–exon organisation of P. alecto RLHs. The intron–exon arrangements of pteropid bat RIG-I, mda5 and LGP2 were predicted by nucleotide alignment of P. alecto mRNA and P. vampyrus genomic DNA. Predicted coding and non-coding exons are drawn as light and dark rectangles respectively, while putative introns are indicated by dotted lines. The figure is drawn to scale and distances are given in bp, however some intron sizes may be underestimated due to gaps in the P. vampyrus genome assembly. mda5 exon 8 is absent from the P. vampyrus genome assembly due to a sequencing gap, however it is expected to be a contiguous exon in the pteropid bats as it is highly conserved and present as a single exon in other species examined. In all other cases, intron–exon boundaries were clearly identified by homology with other species and by the presence of splice site consensus sequences.