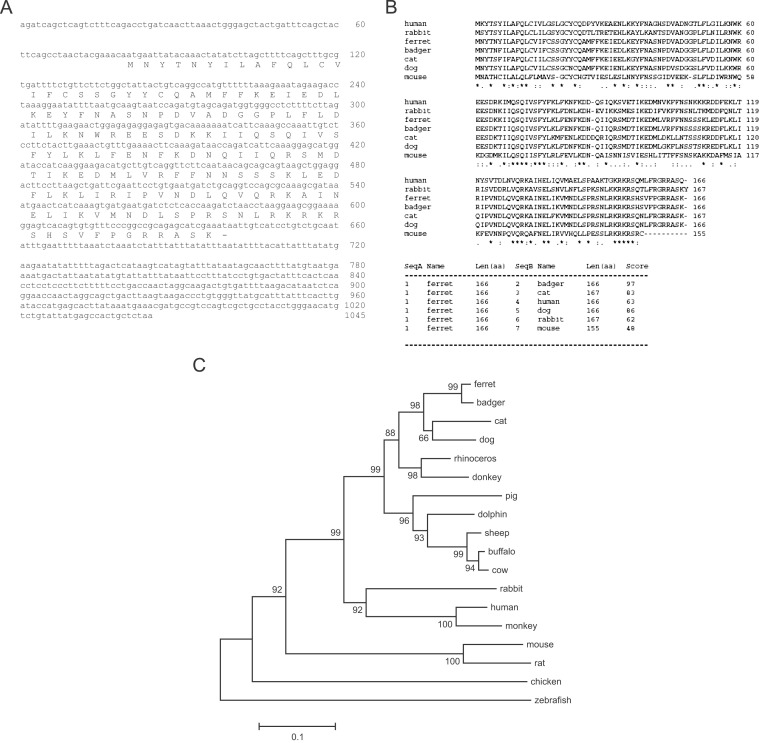
Figure 1.
Ferret IFN-γ cDNA. (A) Full-length ferret IFN-γ cDNA sequence including 80 base pairs in the 5′ untranslated region (UTR), 501 base pairs of coding sequence with predicted amino acid sequence, and 404 base pairs in the 3′UTR. (B) Alignment of the amino acid sequences of ferret, Eurasian badger, rabbit, cat, dog, mouse, and human IFN-γ precursor proteins (accession numbers Y11647, P30123, P46402, P42161, P01580, and P01579, respectively) is shown. Asterisks indicate positions displaying identical amino acid residues in all sequences in the alignment, and periods indicate positions displaying semiconserved substitutions. Scores of amino acid homology between ferret IFN-γ and IFN-γ from different species are shown in the lower panel. (C) Phylogenetic tree showing the relationship between ferret and other known vertebrate IFN-γ sequences. This tree was constructed using CLUSTAL W and MEGA 3.1 packages and bootstrapped 10,000 times. †Bootstrapping confidence values are between 66 and 100. The Gene peptide accession numbers for IFN-γ are: badger, CAA72346; dog, AAD314233; panda, ABE02189; cat, BAA06309; rhinoceros, ABC18310; donkey, AAC42595; pig, ABG56234; dolphin, BAA82042; sheep, ABD64367; buffalo, BAE75855; cow, NP_776511; armadillo, AAZ57195; woodchuck, AAC31963; rabbit, BAA24439; human, P01579; monkey, AAM21477; mouse, P01580; rat, NP_620235; chicken, CAA69227; zebrafish, BAD06253.