Fig. 4.
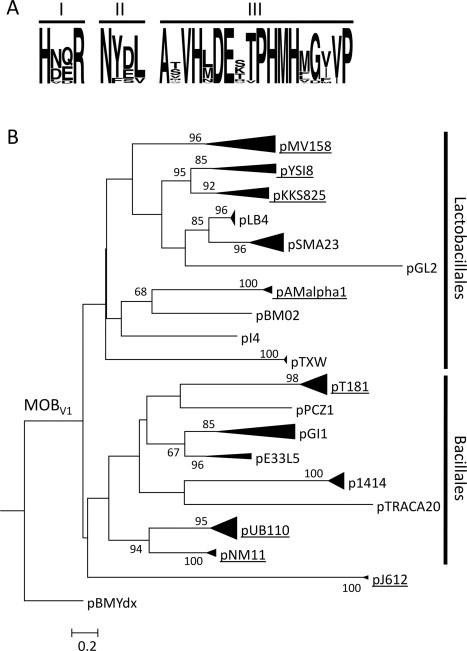
Phylogeny of MobMpMV158 homologs. (A) Logos of the MOBV1 relaxase motifs. MOBV1 relaxases were analyzed using WebLogo (version 2.8.2) (Crooks et al., 2004). (B) The 300 N-terminal residues of the MobM relaxase of plasmid pMV158 were used as query in a PSI-BLAST search (Altschul et al., 1997) (e-value: 1 × E−6 and limited to 100 non-redundant plasmid hits). The search converged in the sixth iteration. The 300 N-terminal residues of the homologs were aligned using MUSCLE (Edgar, 2004). The phylogenetic reconstruction was carried out by maximum likelihood (ML), using RAxML version 7.2.7 (Stamatakis, 2006). 100 ML trees were executed using the JTTGAMMA model. 1000 bootstrap trees were then inferred to obtain the confidence values for each node of the best ML tree. Only bootstrap values >50% are indicated. The MOBV2 relaxase of plasmid pBMYdx (GenBank Acc. No. NP_981974.1) was used as outgroup. Highly related clusters are compressed and a prototype member is indicated. The names and features of all members included in the tree are recorded in Table 1. Clusters grouping plasmids that encode antibiotic resistance traits are underlined. Vertical bars delimit clades for which most of their members are hosted either in Lactobacillales or in Bacillales.
