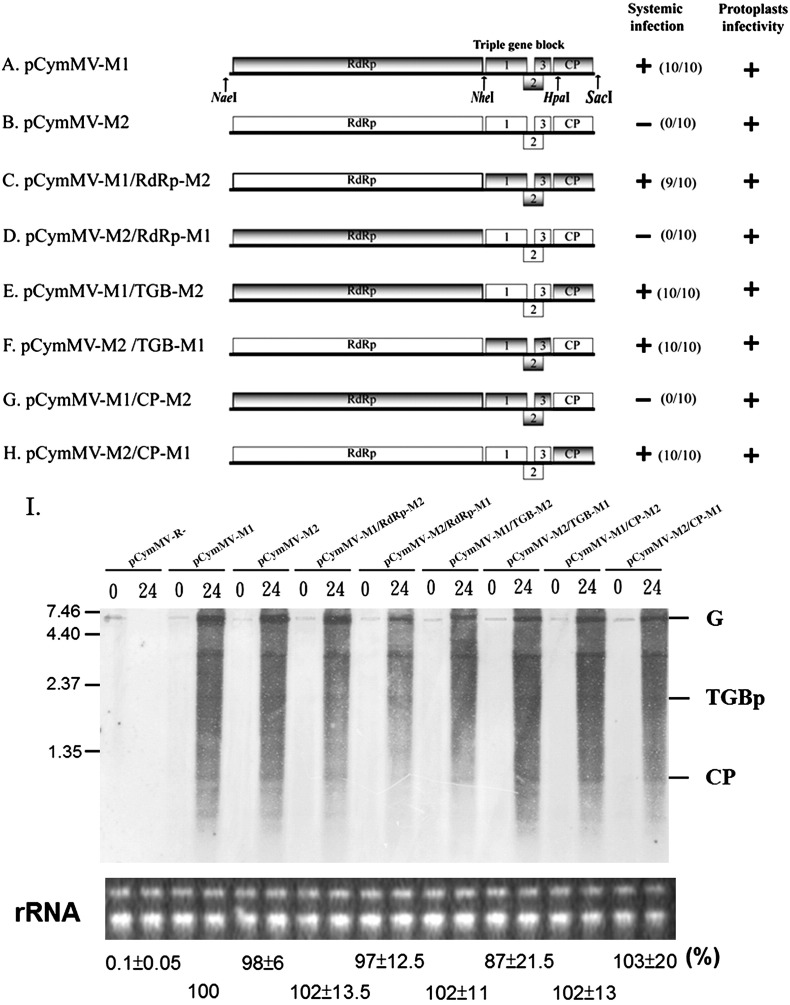
Fig. 3.
Schematic representation of genome organization and infectivity assay of the parental CymMV-M1 and CymMV-M2 and the derived chimeric constructs. (A–H). Rectangles represent open reading frames encoded by CymMV genomic RNA, RNA-dependent RNA polymerase (RdRp), triple gene block (TGB) ORFs 1, 2, and 3 and capsid protein (CP). Sequences corresponding to pCymMV-M1 and pCymMV-M2 are indicated by gray and white rectangles, respectively. The restriction enzyme sites for constructing chimeric viruses are indicated. (Although HpaI sites are located 129 nt downstream of CP translation start sites, the amino acid sequences in the regions between M1 and M2 are identical.) Clones competent in protoplast accumulation and systemic infection in N. benthamiana are indicated by +, and the ratio of systemic infected to total inoculated plants is indicated. Systemic infection was detected 2 weeks post-inoculation by RT-PCR. (I) Protoplast infectivity was detected 24 h post-inoculation by northern blot hybridization, and the ribosomal RNA used for a loading control are indicated. Genomic RNA (G), TGBp, and CP subgenomic RNA are indicated. The pCymMV-R- used as a negative control is illustrated in Fig. 1. The average percentage of relative real-time RT-PCR quantification (from 3 independent experiments) of CymMV RNA from CymMV clone-infected protoplasts at 24 h post-inoculation is indicated. The accumulation of pCymMV-M1 was set at 100% for relative quantification. Numbers at the left correspond to positions of marker RNAs (sizes in 1000 nucleotides) analyzed in the same gel.