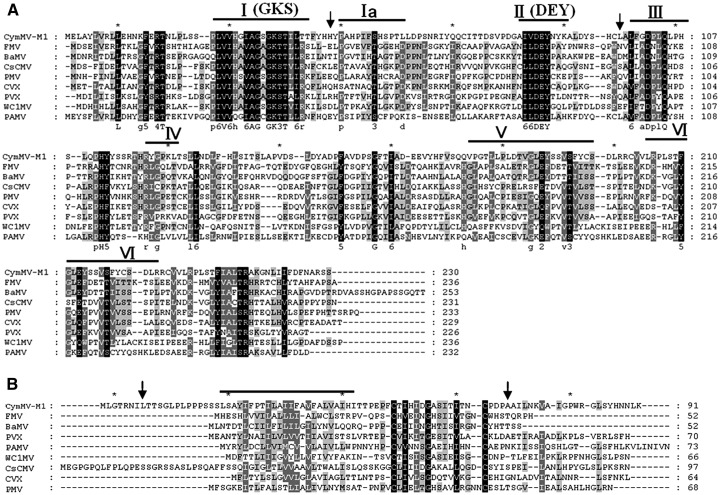
Fig. 9.
The amino acid sequence alignment of TGBp1 NTP/helicase domain and TGBp3 transmembrane domain of potexvirus. The amino acid sequence alignments were conducted by use of clustal X 1.83 (Thompson et al., 1997). (A) The conserved motifs of the potexvirus NTPase/helicase domain were previously predicted (Kalinina et al., 2002). The 7 predicted NTPase/helicase motifs of potexvirus TGBp1 are shown, and two canonical motifs of NTPase, DEY and GKS, are indicated. The arrows indicate the amino acid positions important for pCymMV-M2 to systemically infect N. benthamiana. (B) The potexvirus TGBp3 transmembrane domain was previously identified (Krishnamurthy et al., 2003). The CymMV transmembrane domain predicted by DAS program (http://www.sbc.su.se/~miklos/DAS/maindas.html) is indicated by a thick black line. The arrows indicate the amino acids important for pCymMV-M2 to systemically infect N. benthamiana. The viruses, abbreviation and accession number used in alignments of TGBp1 NTP/helicase and TGbp3 transmembrane domains are described below. Cymbidium mosaic virus (CymMV, accession number AY571289); Cymbidium mosaic virus (CymMV, accession number AY571289); Foxtail mosaic virus (FMV, accession number NC_001483); Watermelon spotted wilt virus (BSMV, accession number NC_003481); Cassava common mosaic virus (CsCMV, accession number NC_001658); Papaya mosaic virus (PMV, accession number NC_001748); Cactus virus X (CVX, accession number NC_002815); Potato virus X (PVX, accession number NC_001455); White clover mosaic virus (WC1MV, accession number X06728).