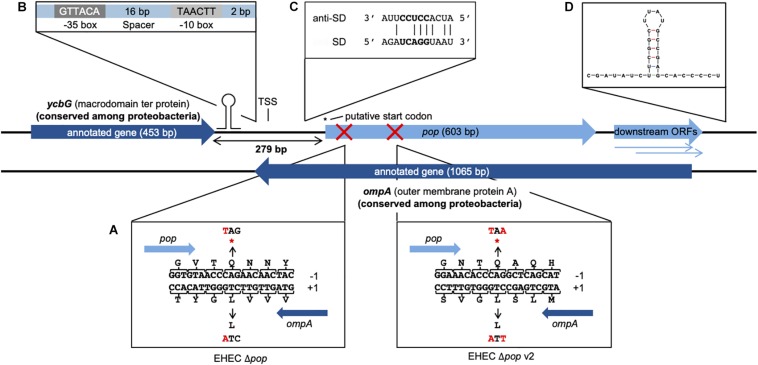
FIGURE 1.
Genomic organization and operon structure of pop. pop (603 bp) is located downstream of the annotated gene ycbG and completely embedded antisense in the sequence of ompA. Downstream of pop exist two smaller overlapping open reading frames, which overlap ompA almost completely and are referred to as downstream ORFs. A transcription start site (TSS) was experimentally identified in the intergenic region of pop and ycbG. The full genomic sequence of pop is given in Supplementary Figure S1. (A) Design of translationally arrested mutants of pop. The overlapping ORF pop is located in reading frame −1 with respect to ompA. Genomic mutants for phenotypic characterization contained a single base substitution C → T at genome position 1236083 for EHEC Δpop or C → T and G → A at genome positions 1236302 and 1236304 for EHEC Δpop v2 (indicated with red crosses), leading to stop codons (*) in pop and synonymous changes in ompA. (B) Promoter sequence. The sequences of the −10 box and −35 box as well as the length of the spacer between the conserved boxes and the distance to the TSS are shown. (C) Alignment of Shine-Dalgarno (SD) sequence of pop and anti-Shine-Dalgarno (anti-SD) sequence. The predicted SD sequence (ΔG° = −3.6 kcal/mol) upstream of the putative start codon is aligned to the consensus of the anti-SD sequence of the 16S rRNA in the 30S ribosomal subunit (Ma et al., 2002). The core of the ribosome binding site is displayed in bold letters. (D) Secondary structure of the first 40 bp of the predicted terminator. The folding was conducted with Mfold and the structure has a final energy of ΔG = −8.6 kcal/mol.