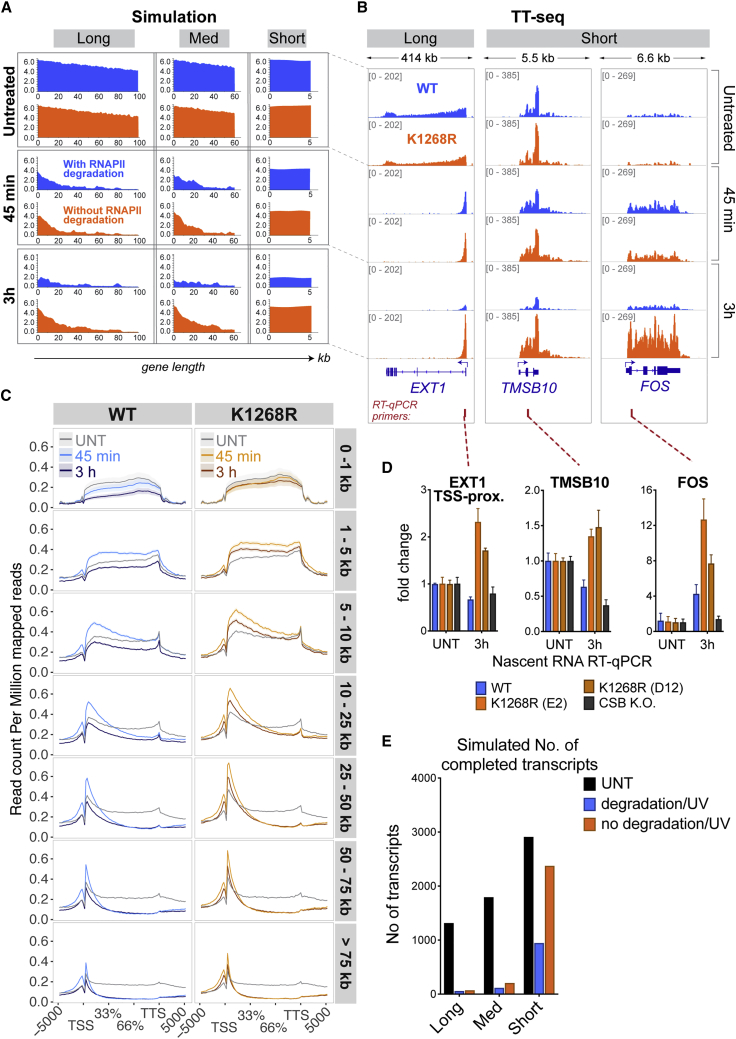
Figure 4.
K1268 Ubiquitylation Ensures that Short Genes Also Cease Expression upon UV Irradiation
(A) Simulated RNAPII activity on a long, medium, and short gene, with or without DNA damage. Three genes are competing for the same pool of RNAPII molecules. The initiation probability was weighted by the relative representation of long (>100 kb), medium (30–100 kb), and short (<30 kb) genes in the genome (0.1: 0.2: 0.7, respectively). RNAPII degradation upon stalling was either allowed (blue) or not (orange).
(B) Browser tracks of TTchem-seq data, from a long (EXT1) and two short genes (TMSB10 and FOS). The data are normalized to yeast spike-in. RT-qPCR primers used for validation are indicated below gene panels.
(C) Metagene TTchem-seq profiles of all genes in the genome, stratified by gene length (indicated in bold on the right). x axis: relative scale (TSS and TTS are indicated); y axis: reads per million mapped reads (rpm). Transcription levels in untreated cells (gray lines), and 45 min (light-colored lines) and 3 h (dark-colored lines) after UV irradiation (20 J/m2) are shown. The data are normalized to yeast spike-in.
(D) Nascent RNA production after UV irradiation (20 J/m2) at TSS-proximal regions of EXT1, TMSB10, and FOS genes. RT-qPCR primer positions are indicated in (B). Data are represented as mean ± SD and normalized to the mature GAPDH transcript and to untreated conditions.
(E) Simulation-predicted number of mRNA transcripts in a long, medium, and short gene, in untreated cells and 4 h post-damage, in scenarios where RNAPII degradation is allowed (WT equivalent) or not (K1268R equivalent). Parameter values as in (A).