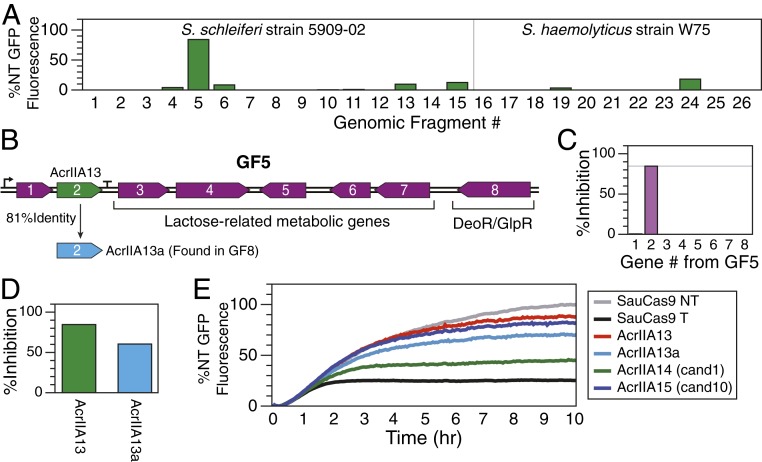
Fig. 2.
Identification of three Acrs using amplicon screening and guilt by association. (A) The relative level of S. schleiferi (Left) or S. haemolyticus (Right) Cas9 DNA cleavage inhibition for each fragment was measured as percentage of the GFP expression for the nontargeting (NT) control after subtracting the fluorescence level observed in the targeting (T) control with no Acr present. Thus, 0% inhibition is equivalent to the GFP expression level measured with the targeting sgRNA, while 100% inhibition represents no reduction in the maximum GFP expression. GF5 is the only amplicon that exhibits Acr activity. (B) Genes found in GF5. (C) Each gene in GF5 was individually cloned and tested for Acr activity with TXTL and SauCas9. The second gene of GF5 (AcrIIA13) inhibits SauCas9. (D) AcrIIA13a, a homolog of AcrIIA13, is found in GF8 and is also able to inhibit SauCas9 in a TXTL assay. GF8, which contains AcrIIA13a, did not exhibit Acr activity, however. (E) Of 10 candidates chosen from a guilt-by-association search seeded by GF5 gene 1, candidates 1 (AcrIIA14) and 10 (AcrIIA15) were found to inhibit SauCas9 using the TXTL assay. All data in this figure are from single TXTL runs.