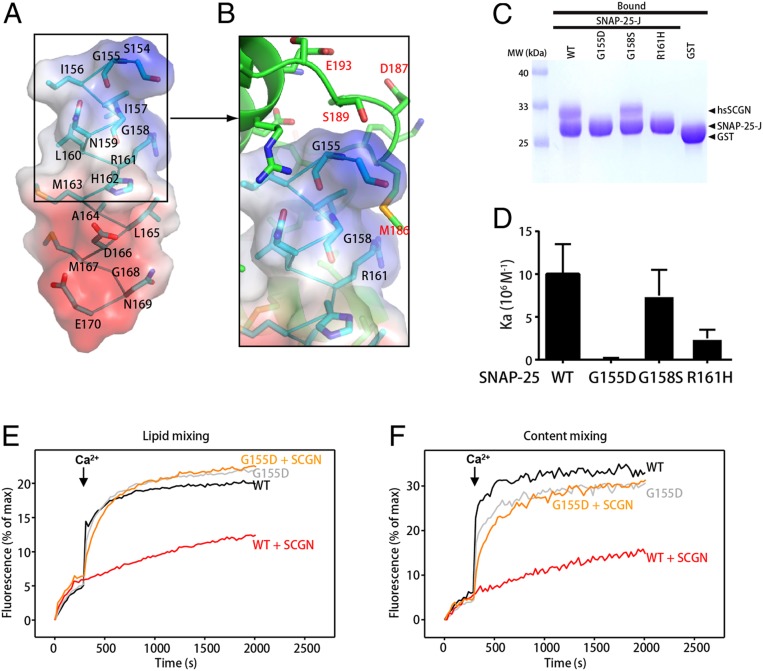
Fig. 4.
Identification of SNAP-25 residues that specifically interact with SCGN. (A) Structure of the SNAP-25 peptide (stick presentation) observed in the SCGN–SNAP-25 complex, together with its electrostatic potential surface. Blue: positive potential; red: negative potential. (B) Detailed view of the interactions involving drSCGN and the N terminus of the SNAP-25 peptide. Three SNAP-25 residues (G155, G158, R161) that were mutated in later studies were highlighted. (C) GST pull-down assays performed with GST-SNAP-25 J wild-type or mutants, or GST, and purified hsSCGN, in the presence of 2 mM CaCl2 and 0.5% Triton X-100. After incubation with soluble proteins, the resin was extensively washed. The resin-bound proteins were then subjected to SDS/PAGE and Coomassie blue staining. (D) Affinity between SNAP-25 J fragment wild-type or mutants, and human SCGN (hsSCGN) in the presence of 2 mM CaCl2, determined by ITC. Association constants (Ka) were measured from three independent titrations, and shown as mean ± SD. (E and F) Lipid mixing (E) between V- and T-liposomes was monitored from the fluorescence de-quenching of Marina blue lipids, and content mixing (F) was monitored from the increase in the fluorescence signal of Cy5-streptavidin trapped in the V-liposomes caused by FRET with PhycoE-biotin trapped in the T-liposomes upon liposome fusion. Assays were performed with V- and T-liposomes in the presence of Munc18-1, M13C1C2BMUNC2C, NSF, and αSNAP, with or without 3 μM SCGN. The T-liposomes contained WT or G155D SNAP-25. Experiments were started in the presence of 100 μM EGTA and 5 μM streptavidin, and Ca2+ (600 μM) was added at 300 s.