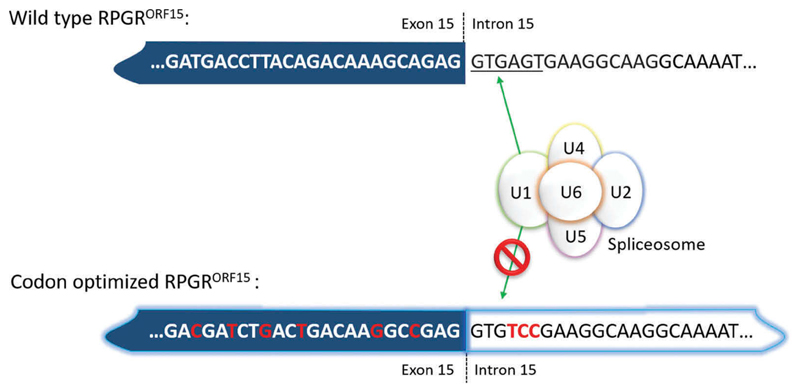
Figure 4.
Codon optimization allows the removal of cryptic splice sites by replacing nucleotides that can be recognized by the spliceosome by other nucleotides without affecting the protein sequence. Hence, this approach makes the nucleotide sequence more stable and less prone to mutations. Note, that in this case, the canonical splice donor site ‘GT … ’ cannot easily be codon optimized because it is in-frame for valine in the ORF15 variant and all four valine amino acid codons have GT in positions 1 and 2. Positions 4–6 in the splice site, however, can be changed completely because they encode serine, which is AGT in the wild-type sequence (fulfilling the splice donor sequence) but TCC in the codon-optimized sequence. Serine has six different codons which provide a wider range of alternate nucleotide sequences to completely disrupt the splice donor site signal, whilst preserving the amino acid sequence in the protein.