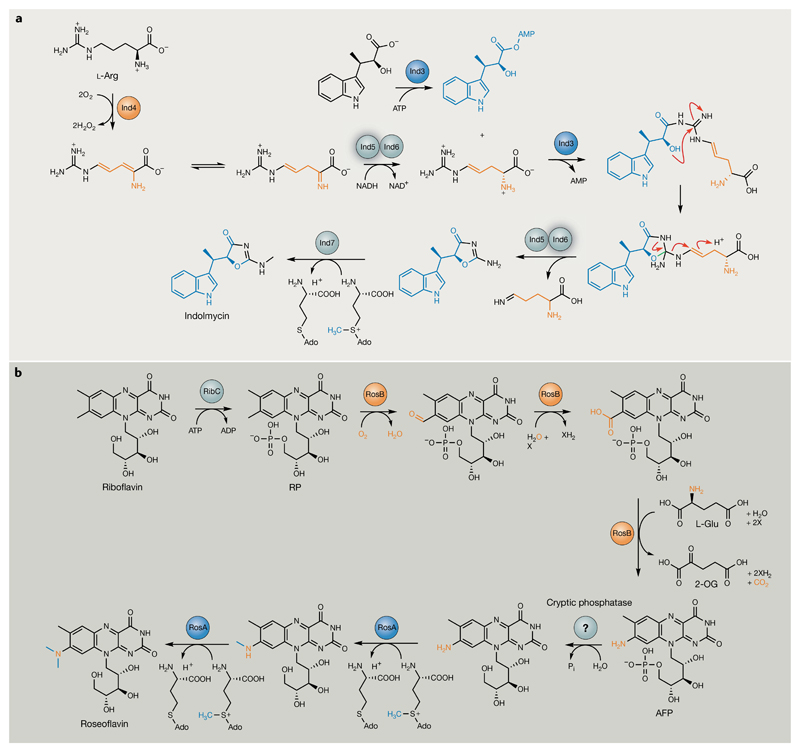
Fig. 10. Unusual enzymology from natural product pathways that lack signature biosynthetic genes.
a | Biosynthesis of indolmycin from l-Arg and indolmycenic acid. Following Ind4 oxidation of l-Arg, the resulting unstable imine product is selectively reduced by the d-specific, NADH-dependent reductase Ind5, preventing off-pathway reactions, such as deamination, from occurring190. Ind3 catalyses an ATP-dependent condensation of d-4,5-dehydroarginine with indolmycenic acid, resulting in the formation of the indolmycin oxazoline ring (new bond highlighted in red)189. Ind6 (embedded in a complex with Ind5) performs an unusual gatekeeping role to ensure release of the correct leaving group, again to prevent the generation of off-pathway products. Ind5 and Ind6 are highlighted for their respective transformations. The N-methyltransferase Ind7 completes biosynthesis. b | Roseoflavin biosynthesis from riboflavin196. The ATP-dependent flavokinase RibC first catalyses riboflavin-5′-phosphate (RP) formation. RosB (orange) performs the subsequent three transformations to generate 8-demethyl-8-amino-riboflavin-5′-phosphate (AFP) in the presence of thiamine and l-Glu, with 2-oxoglutarate (2-OG) produced as a by-product195,196. The RibC-installed phosphate group is subsequently removed by a cryptic phosphatase, before sequential N-methylations catalysed by the S-adenosyl methionine (SAM)-dependent dimethyltransferase RosA194 (blue). X represents a cryptic hydrogen acceptor. Modifications catalysed by RosA and RosB have been highlighted in their respective colours.