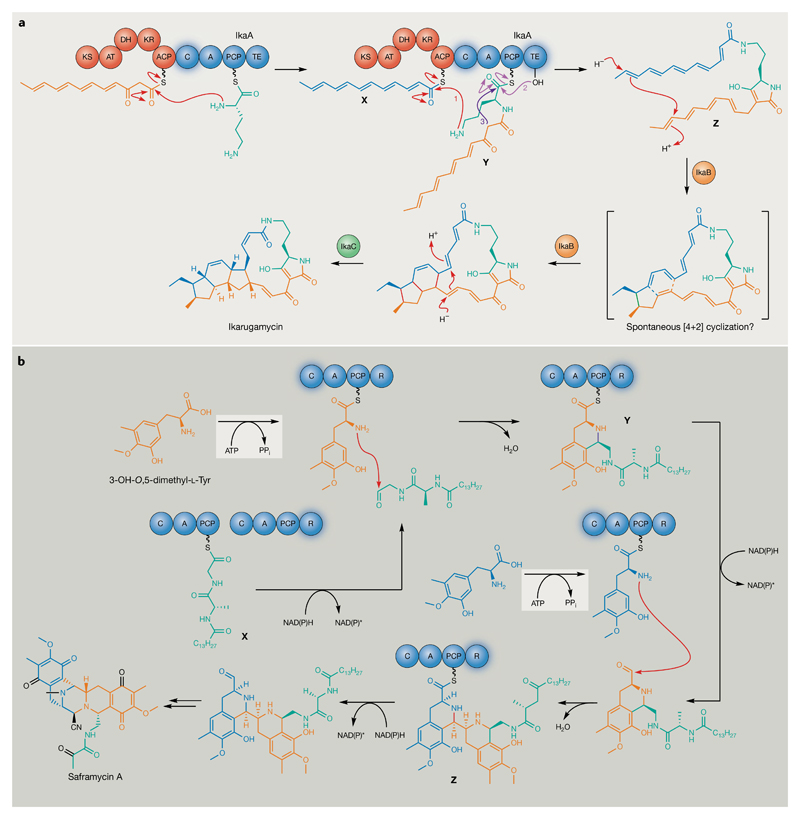
Fig. 5. Polyketide synthase-catalysed and nonribosomal peptide synthetase-catalysed transformations that define novel natural product families.
a | Members of the polycyclic tetramate macrolactam family of polyketides are produced by the activities of only three enzymes, as illustrated for ikarugamycin72–74. New bonds formed during successive cyclizations are highlighted in red. Steps 1–3 illustrate amide bond formation between thioester intermediates X and Y by the IkaA C domain (step 1), transfer of the resulting thioester intermediate to the IkaA thioesterase domain active site Ser residue (step 2) and thioesterase-catalysed intramolecular attack to yield the pyrroline moiety with concomitant release of intermediate Z (step 3). It is not clear whether formation of the six-membered ring is spontaneous or enzyme-catalysed. The timing and nature of the isomerization event that results in the introduction of a cis double-bond in the final ikarugamycin pathway product is also not clear; however, the IkaC homologue OX4 is proposed to be responsible for cis double-bond introduction during the biosynthesis of heat-stable antifungal factor, a polycyclic tetramate macrolactam produced by Lysobacter enzymogenes226–228. b | A single nonribosomal peptide synthetase module catalyses iterative Pictet–Spengler-type cyclizations and reductions of structurally different intermediates to assemble the characteristic scaffold of the tetrahydroisoquinoline antibiotics76. Structurally different SfmC reductase (R) domain peptidyl thioester intermediates are indicated by X–Z. Adenylations of 3-OH-O,5-dimethyl-l-Tyr residues before condensation (C)-domain-catalysed cyclization by the SfmC adenylation (A) domain are indicated in grey. New bonds formed by the C domain are highlighted in purple. Individual substrates are highlighted in different colours so that their fate can be tracked through the mechanism illustrated. ACP, acyl carrier protein; AT, acyltransferase; DH, dehydratase; KR, ketoreductase; KS, ketosynthase; PCP, peptidyl carrier protein; TE, thioesterase.