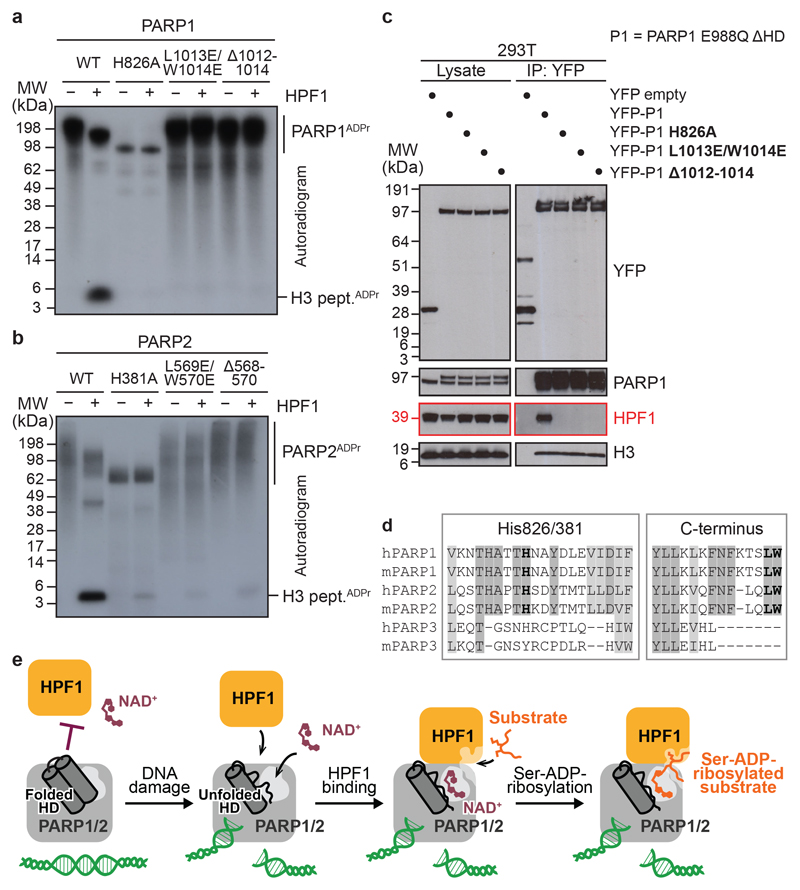
Fig 4. HPF1-interacting PARP1/2 residues and model of DNA damage-induced ADP-ribosylation.
a, b, Radioactive ADP-ribosylation assay of PARP1 or PARP2 mutants +/− HPF1.
c, PARP1 co-immunoprecipitation (IP) from 293T cells treated with olaparib and H2O2.
d, Fragments of a multiple-sequence alignment of human (h) and mouse (m) PARP1, PARP2, and PARP3. Invariant (dark grey) and highly conserved (light grey) residues across at least four of the analysed proteins are highlighted. His826/381 (human PARP1/2 numbering) and the extreme C-terminal Leu-Trp motif are shown in bold.
e, Proposed model of HPF1-PARP1/2-dependent ADP-ribosylation upon DNA damage. The inhibition of HPF1 and NAD+ binding to PARP1/2 is relieved upon PARP1/2 binding to DNA breaks, leading to the formation of a composite HPF1-PARP active site capable of recruiting and modifying KS (shown) or other serine-based substrate motifs.
Experiments in a-c were performed independently three times with similar results.