Figure 6.
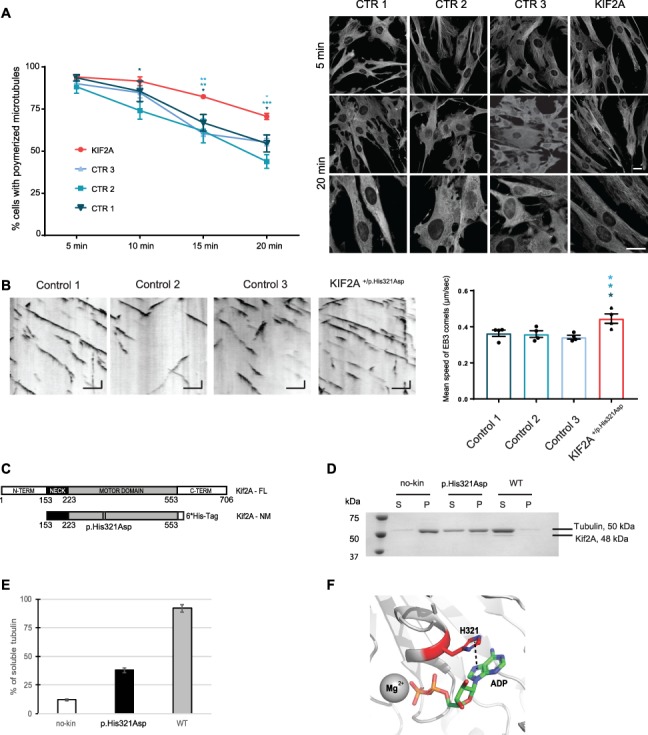
KIF2A His321Asp variant effects on microtubule stability and dynamics. (a) Depolymerization assay in fibroblasts derived from a subject bearing the KIF2A His321Asp variant and three controls. Left panel: Graph represents the percentage of cells with polymerized microtubules over 20 min time after nocodazole treatment. Data are presented as mean ± SEM, n = 3 biologically independent experiments. Two way ANOVA with Tukey’s post hoc test, *P < 0.05, **P < 0.01, ***P < 0.001, compared to controls. Right: representative images of cells at 5 and 20 min of nocodazole treatment, after immunofluorescence staining of tubulin (gray), scale bar 20 μm. (B) Microtubule dynamics in fibroblasts derived from a subject bearing the heterozygous KIF2A p.His321Asp variant and three controls. Left panel: Representative kymographs of dynamic EB3 comets in different controls and mutant fibroblasts. Kymographs are calibrated in time (y, s) and space (x, μm). Scale bar Y = 10 s, X = 5 μm. Right panel: Quantification of EB3 mean velocity. Data are presented as mean ± SEM, n = 4 biologically independent experiments, one-way ANOVA with Dunnett’s post hoc test, *P < 0.05 compared to controls. (C) Biochemical analysis of WT and p.His321Asp mutant of Kif2A. (C) Schematic representation of domain composition of human Kif2A and its constructs used in this study. Location of the H321D mutation is shown. (D) MT depolymerization activity of WT and H321D Kif2A. Taxol-stabilized MTs (1.5 μM) were incubated with WT, H321D (0.075 μM), and no kinesin for 10 min at 25°C. Soluble tubulin and MT polymers were separated into supernatant (S) and pellet (P) by sedimentation. Equal volume of fractions was analyzed on SDS-PAGE gel. (E) Percentage of soluble tubulin in each depolymerization reactions was calculated by densitometry of the bands using ImageJ software. (F) View of the nucleotide-binding site of Kif2A (PDB ID 2GRY). H321 residue (red, sticks) and its pi-interaction with the adenosine moiety of the nucleotide (dash line) are shown. Figure was generated with PyMOL.
