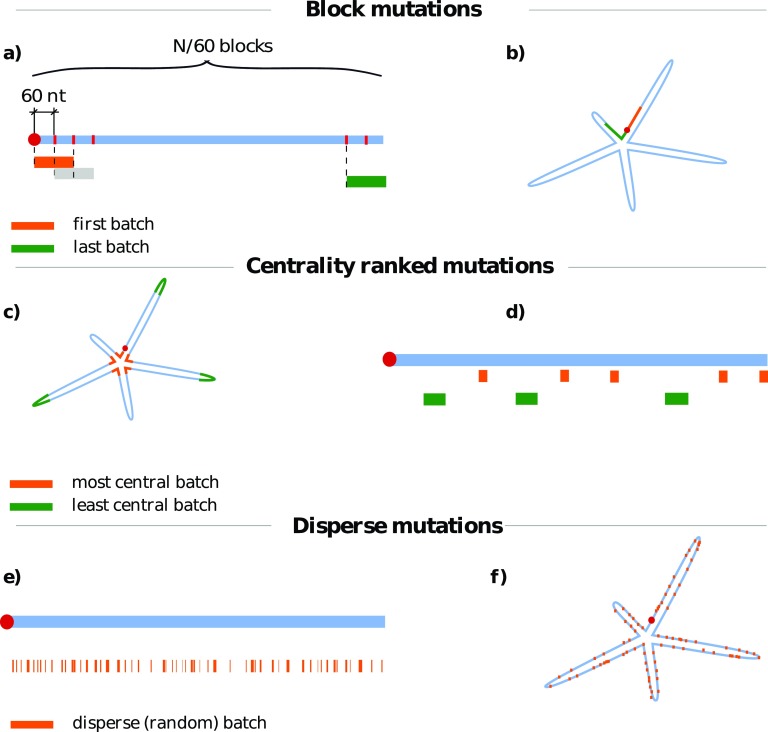
Figure 2.
(a) and (b) Block, (c) and (d) centrality-ranked, and (e) and (f) disperse mutation schemes. For each mutation scheme, a simple sketch of a linear genomic sequence is shown in blue with different mutation batches denoted below it; each color denotes a separate batch. We also show a sketch of an ideal, floret-like fold with five branches stemming from a common center. (a) and (b) Block mutations are performed by mutating the nucleotides of a sequence localized in a contiguous batch of length s (in this case, s = 120 nt). The starting points of different batches then slide along the genome in regular intervals. Block mutations are, for the most part, located on only one strand of the folded RNA duplex. (c) and (d) Centrality-ranked mutations are performed in a similar manner, but the nucleotides are grouped in batches according to their (canonically averaged) distance from the center of the fold. This means that the batch of nucleotides with the lowest distances from the center (orange) will correspond to non-contiguous regions on the genome sequence (d), but will be spatially localized near the center of the average fold (c). The batch of nucleotides with the largest distance is shown in green. (e) and (f) Disperse mutations are obtained by randomly assigning nucleotides to batches of a certain size. These mutations show no localization on either sequence (e) or fold (f).