Figure 1.
Loss of HE-Mediated Receptor Binding in Human Betacoronaviruses
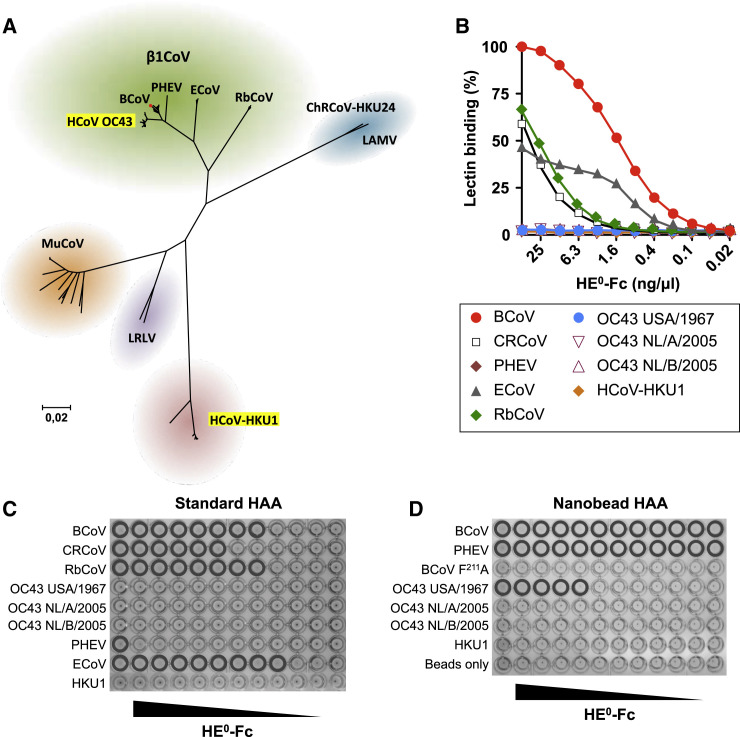
(A) Evolutionary relationships among lineage A betacoronaviruses. Neighbor-joining tree based on βCoV lineage A ORF1b sequences in the NCBI database (n = 206), with 100% bootstrap support for all major branches. Evolutionary distances were computed using the Maximum Composite Likelihood method in MEGA6 (Tamura et al., 2013). The positions of human coronaviruses OC43 and HKU1 are highlighted relative to those of animal lineage A betacoronaviruses, including the various β1CoV subspecies and classical mouse hepatitis virus-type murine coronavirus (MuCoV). ChRCoV-HKU24, Chinese rat coronavirus HKU24 (Lau et al., 2015); LAMV, Longquan Aa mouse coronavirus (Wang et al., 2015); LRLV, Longquan RI rat coronavirus (Wang et al., 2015). The position of CRCoV, a recent split-off of BCoV, is indicated by a red dot. (See also Figure S1.)
(B) HE0-Fc lectins (2-fold serial dilutions, starting at 50 ng/μL) were compared by sp-LBA for relative binding to BSM (with 50 ng/μL BCoV-HE0-Fc set at 100%).
(C) Conventional HAA with 2-fold serial dilutions of HE0-Fc fusion proteins of β1CoV members and HCoV-HKU1 (starting at 25 ng/well). Wells positive for hemagglutination are encircled.
(D) High-sensitivity nanobead HAA. Non-complexed nanobeads (“beads only”) and nanobeads complexed with lectin-inactive mutant BCoV HE0 F211A were included as negative controls.