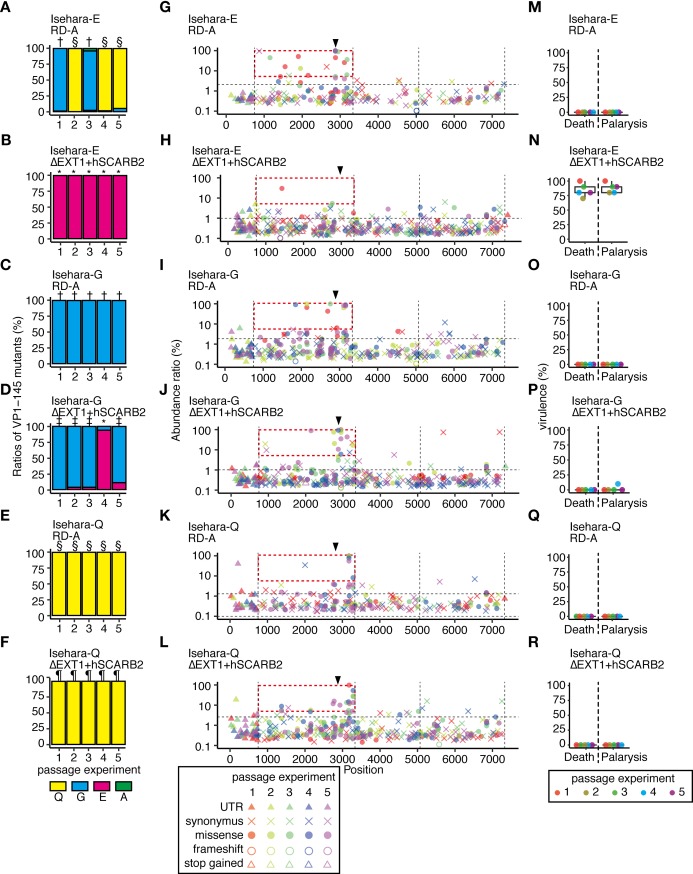
Fig 5. Genome stability and virulence in EV71 populations with or without VP1-145 mutation that were prepared from cDNA clones.
Viruses recovered from transfection of RD-A or ΔEXT1+hSCARB2 cells with in vitro-transcribed RNA from infectious cDNA of Isehara-E, -G, and -Q were passaged in these cells three times. (A–F) The abundance of VP1-145 mutants in the virus population before and after passage, estimated from NGS data using ShoRAH software. VP1-145E-dominant populations resulting from three passages in ΔEXT1+hSCARB2 cells, VP1-145G-dominant populations passaged in RD-A cells, VP1-145G-dominant populations passaged in ΔEXT1+hSCARB2 cells, VP1-145Q-dominant populations passaged in RD-A cells, and VP1-145Q-dominant populations passaged in ΔEXT1+hSCARB2 (analyzed in Figs 6 and 7 and Table 2) are indicated with *, †, ‡, §, and¶, respectively. (G–L) To detect SNVs in passaged virus populations, NGS reads were mapped to the sequence of the cloned cDNA and analyzed using LoFreq and SnpEff software. The abundance of each annotated SNV was plotted on the corresponding position in the virus genome. Vertical dotted lines indicate borders separating genetic regions (5’UTR, P1, P2, P3, and 3’UTR). Horizontal dotted lines indicate the 1.5 IQR of the abundance of each SNV mapped to the graph. Arrowheads indicate the position corresponding to VP1-145. High frequency (>5%) mutations in the P1 region are boxed with a red dashed line. (M–R) Passaged viruses were used to infect hSCARB2 tg mice (105 TCID50, ip). Paralysis and death of the infected mice were observed over 14 days.