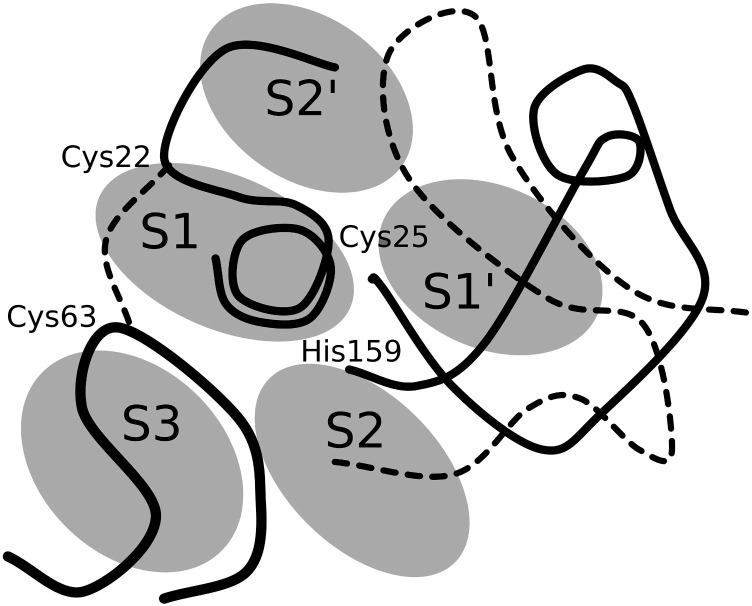
Fig. 1.
3D schematic representation of the substrate-binding sites of papain-like proteases along the active-site cleft. The representation is based on the proposed revised definition of substrate-binding sites based on the crystal structures of substrate-mimicking inhibitors bound to the protease's active sites [11]. The substrate-binding sites of the papain-like proteases are located on the left (S1, S3, S2′) and right (S2, S1′) side of the active-site cleft in accordance with the standard view orientation. According to papain numbering, L-domain loops include residues Gln19-Cys25 and Arg59-Tyr67 whereas R-domain loops contain residues Leu134-His159 and Asn175-Ser205, respectively. The active-site residues Cys25 and His159 and the disulphide Cys22-Cys63 are indicated.