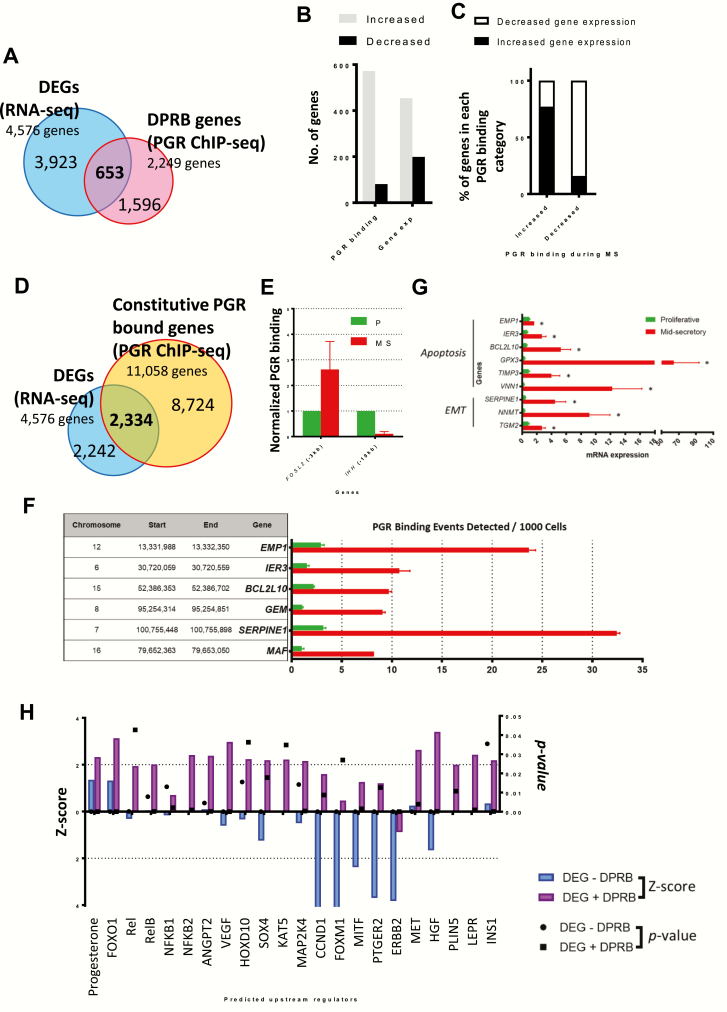
Figure 3.
Identification of Progesterone Receptor (PGR)-regulated genes during the menstrual cycle. (A) Overlaying the genes with differential PGR bound (DPRB) and differential expression identified 653 genes during the P to MS transition. (B) Number of genes showing increased and decreased PGR binding and expression in the endometrium during MS. (C) The percentage of genes showing increased or decreased expression with increased or decreased PGR binding from P to MS. (D) Overlaying the genes with proximal constitutive and PGR binding and differential expression identified 2334 such genes during the P to MS transition. (E) PGR binding activity near two known target genes, FOSL2 and IHH were examined by PGR ChIP-qPCR to confirm the phases of endometrial sample from which chromatin was obtained. qPCR was conducted in triplicates for each sample, and results shown are normalized to values from the P phase, n = 2 independent patients. (F) PGR occupancy was validated for selected genes from the xenobiotic metabolism, apoptosis and epithelial-mesenchymal transition (EMT) pathways using ChIP-qPCR. Experiments were performed using two sets of paired patient samples (each consisting of one P and one MS), and a representative result is shown. *P < 0.05. (G) Selected genes from the xenobiotic metabolism, apoptosis and EMT pathways were validated using qPCR, n = 6 and * P < 0.05. (H) Comparison of the upstream regulator activity (as indicated by the Z-score) for DEGs with and without differential PGR binding. Activity status (Z-score) is plotted on the left Y-axis (blue and purple bars, representing without DPRB and with DPRB, respectively), and significance (p value) is plotted on the right Y-axis (circle and square, representing without DPRB and with DPRB, respectively).