Figure 1.
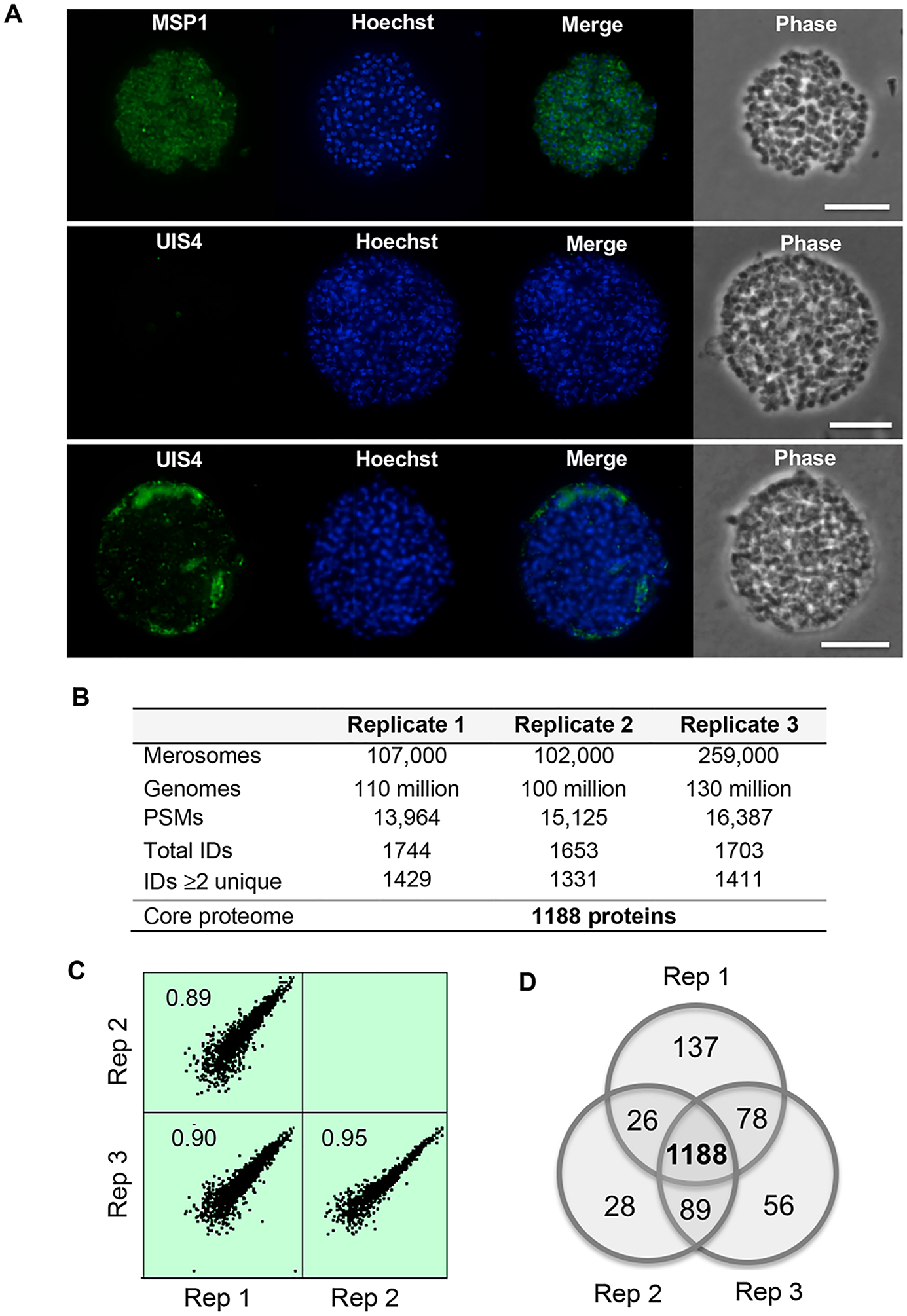
Overview of the core P. berghei merosome proteome. (A) Immunofluorescence analysis of merosome samples used for proteomics. Top: Staining with antibodies specific for MSP1 identifies hepatic merozoites within merosomes and confirms merosomes are intact. Middle and bottom: Staining with antibodies specific for the PV marker UIS4 demonstrates the vacuole is absent or fragmented in merosomes. DNA stained with Hoechst. Scale bar 10 μm. (B) Overview of the P. berghei merosome proteome. Genomes refer to the number of hepatic merozoite genomes estimated by quantitative PCR. PSMs refers to peptide spectrum matches. Total IDs are proteins identified by at least one peptide. IDs ≥ 2 unique are proteins identified by at least two unique peptides. (C) Correlation among biological replicates. iBAQ values were log2 transformed to generate scatter plots showing the correlation among biological replicates. The Pearson correlation between replicates is indicated in each plot. Data shown are for proteins with >2 unique peptides and plots were generated using Perseus version 1.6.0.2078. (D) Concordance between protein IDs with ≥2 unique peptides between replicates. We define the core merosome proteome as the 1188 proteins identified by ≥2 unique peptides in all three biological replicates.
