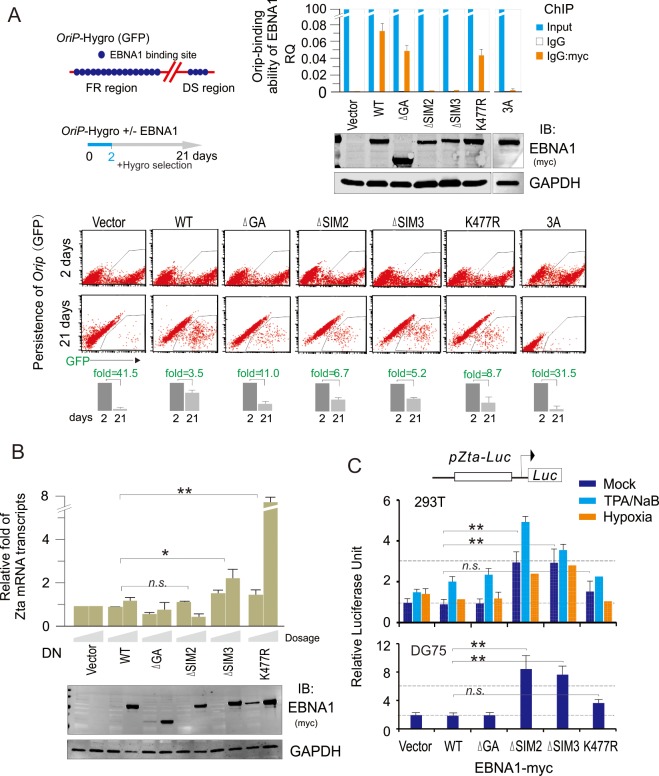
Fig 4. The EBNA1SIM motif is important for DNA binding and maintenance of EBV OriP DNA.
(A) Deletion of EBNA1SIM reduces the DNA-binding ability of EBNA1 and the capacity of EBNA1-mediated OriP maintenance. HEK293 cells were transfected with OriP-GFP-Hygromycin plasmid together with vector encoding the WT or different mutants of myc-tagged EBNA1 or vector alone as indicated. The transfection efficiency was monitored by GFP expression. At 48 h post-transfection, cells were subjected to ChIP assays with normal mouse IgG or mouse anti-myc (9E10) antibodies followed by real-time quantitative PCR with OriP primers (upper panel). The level of OriP per transfection was compared to vector alone and shown as input. The results of OriP-binding ability of EBNA1 are presented after normalization with the protein levels of EBNA1. The expression of WT EBNA1 and its mutants were evaluated by IB assay and are shown in the figure (right panels). For flow-cytometry assays of EBNA1-mediated OriP maintenance, ten thousand of transfected cells were seeded at 2 days post-transfection and selected with 100 μg/ml Hygromycin for 21 days (bottom panels). The average of GFP-positive cells quantified from duplicate experiments is shown in the figure. (B) Dominant negative (DN) mutant of EBNA with SIM deletion reactivates EBV lytic replication. HEK293/Bac-EBV-GFP stable cells were transfected with different amounts of expression plasmids as indicated in the figure. At 48 h post-transfection, the relative level of Zta mRNA transcripts in transfected cells was detected by quantitative PCR. The expression levels of WT EBNA1 and its mutants were evaluated by IB assay and shown in the bottom panel. (C) Reporter assays of the Zta promoter in the presence of EBNA1 under different conditions. HEK293T or DG75 cells were individually co-transfected with a Zta promoter-driven Luciferase reporter (pZta-Luc) and vectors encoding the WT EBNA1-myc or its mutants (dGA, SIM2, SIM3, and K477R). At 24 h post-transfection, cells were exposed with 0.2% oxygen, TPA and sodium butyrate (NaB), or untreated as a control for overnight and then harvest for reporter assays. The results were presented by the RLU (relative luciferase unit) fold compared to pZta-Luc with vector alone. Data are presented as means±SD of three independent experiments. **p<0.01, *p<0.05; n.s. no significant.