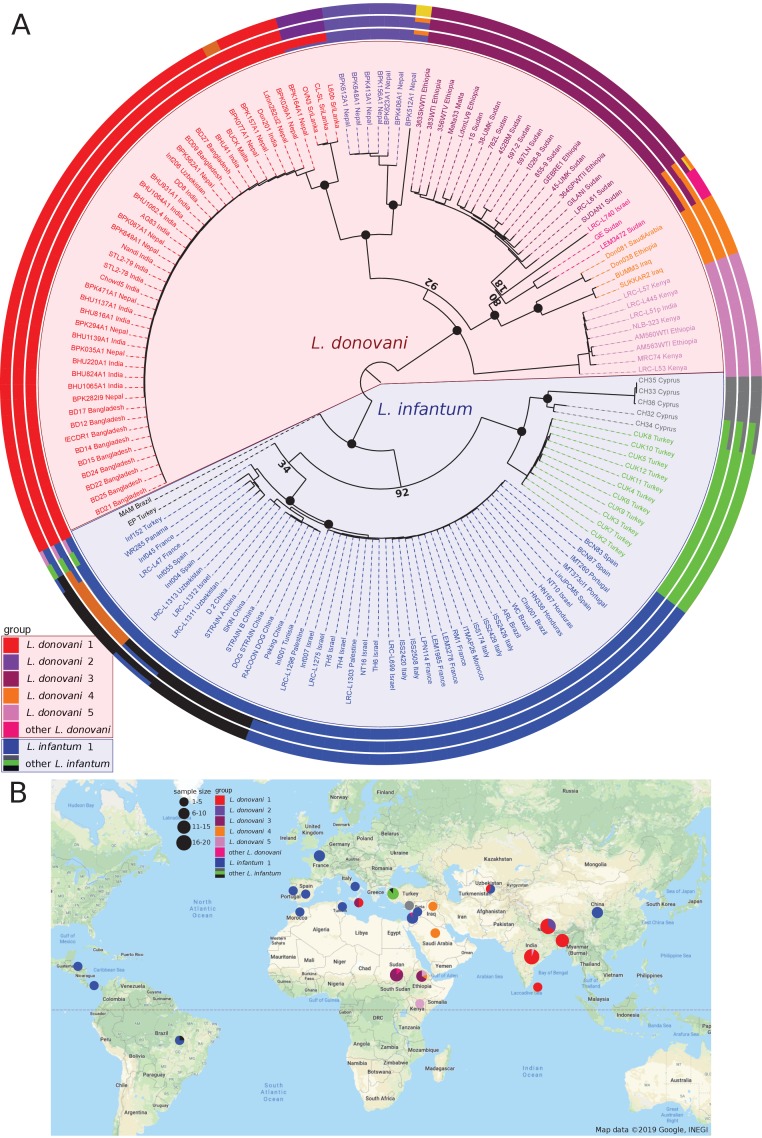
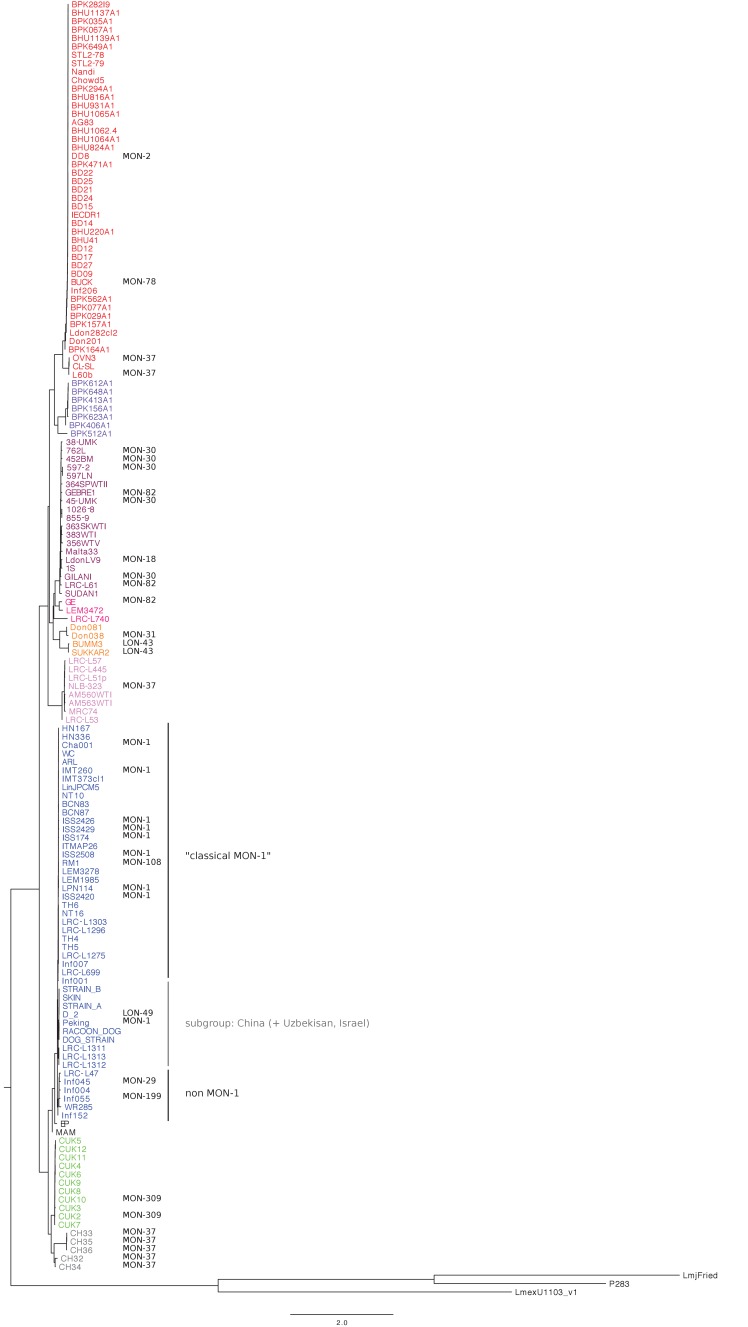
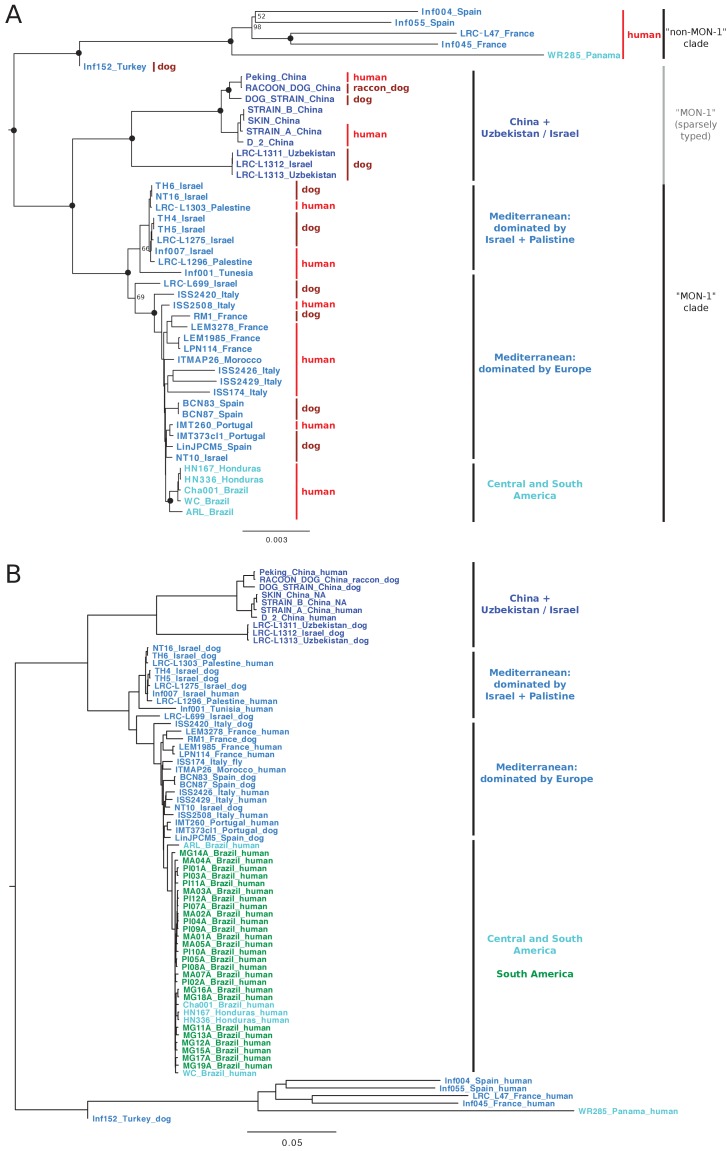
Figure 1. Sample phylogeny and distribution.
(A) Phylogeny of all 151 samples of the L. donovani complex. The phylogeny was calculated with neighbour joining based on Nei's distances using genome-wide SNPs and rooted based on the inclusion of isolates of L. mexicana (U1103.v1), L. tropica (P283) and L. major (LmjFried) (outgroups not shown in the phylogeny). Bootstrap support is shown for prominent nodes in the phylogeny as black circles for values of 100% and otherwise the respective support value in % based on 1000 replicates. The groupings shown in the outer circles were calculated by admixture with K = 8, K = 11 and K = 13 (see Materials and methods). Groups labelled with different colours were defined based on the phylogeny and include monophyletic groups as well as groups that are polyphyletic and/or largely influenced by hybridisation (indicated by ‘other’). (B) Map of the sampling locations. Groups are indicated by the different colours. Sample sizes by country of origin are visualised by the sizes of the circles.