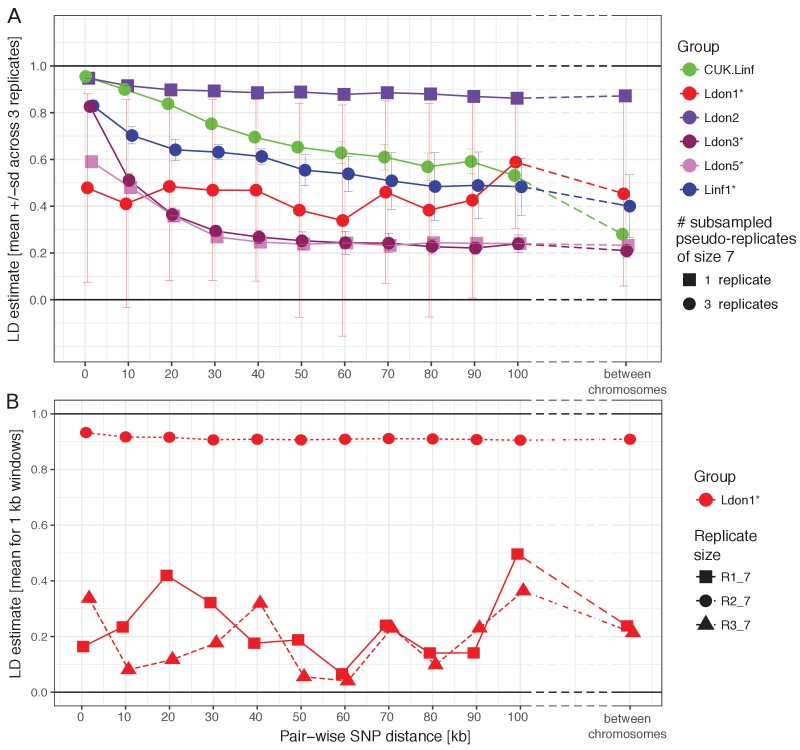
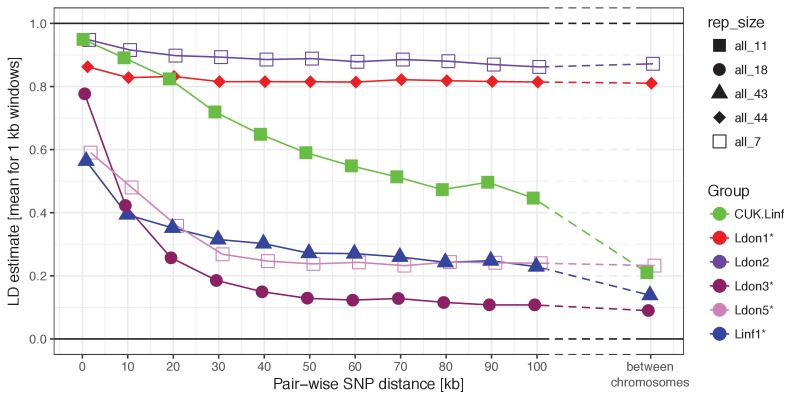
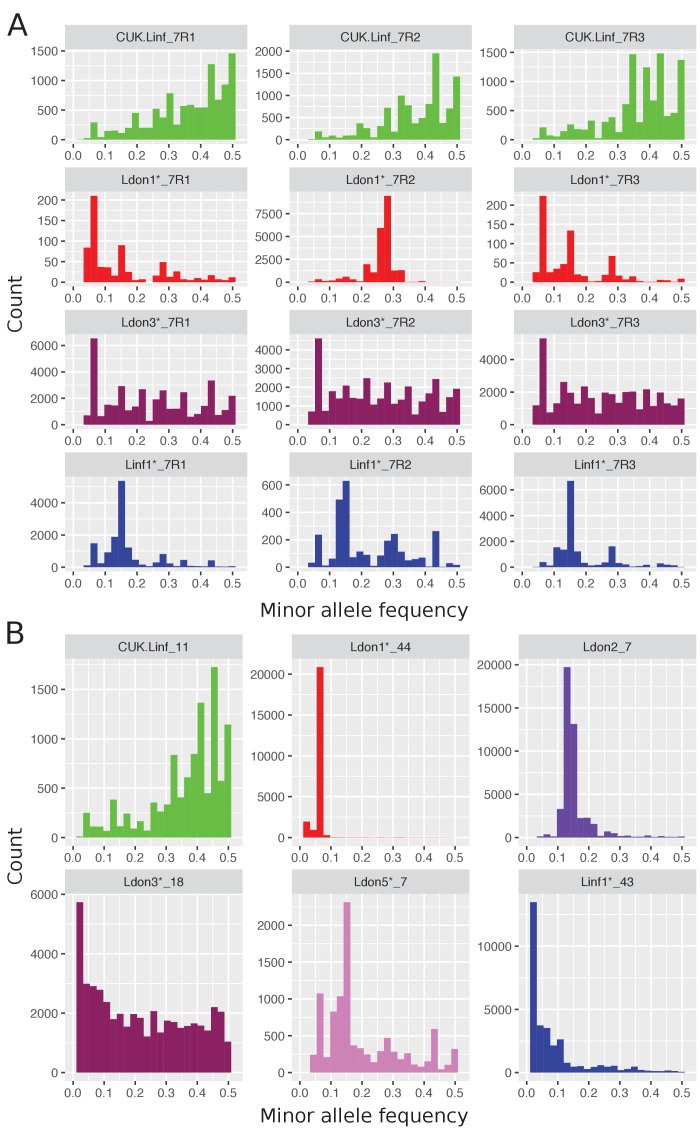
Figure 5. LD decay with genomic distance.
(A) LD decay was measured for the six largest groups removing isolates that were identified as putative strain mixtures (indicated by *; see Materials and methods). Groups with more than seven isolates per group were sub-sampled to three pseudo-replicates of seven isolates (round symbols) to make LD estimates comparable between groups. Mean and standard deviation across the three pseudo-replicates are shown where applicable. Groups with only seven isolates were not sub-sampled and are indicated by squared symbols. (B) LD decay with distance is shown for the three pseudo-replicates for the Ldon1 group. (A and B) Data for individual replicates was calculated as means of 1 kb windows for SNP pairs of the stated genomic distance. For LD estimates between chromosomes, 100 SNPs were randomly sampled per chromosome and means across all pair-wise combinations between chromosomes are shown. This procedure was done twice independently but as differences between both such replicates were negligible, only the results of one replicate are shown.