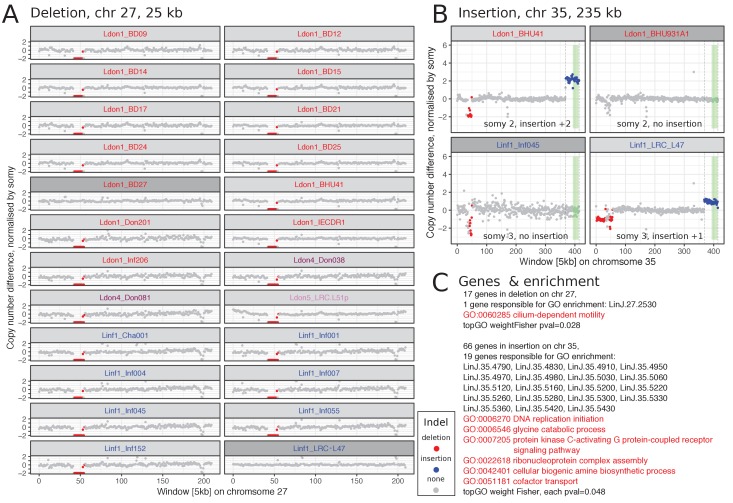
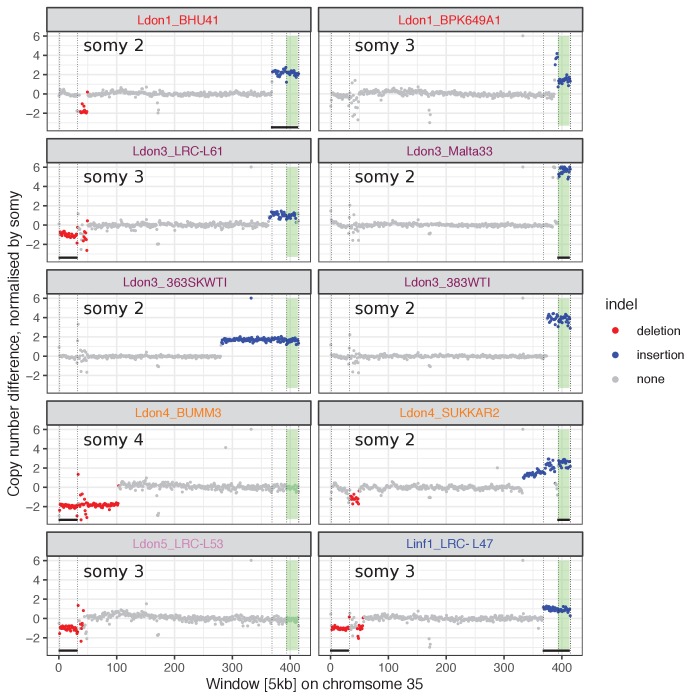
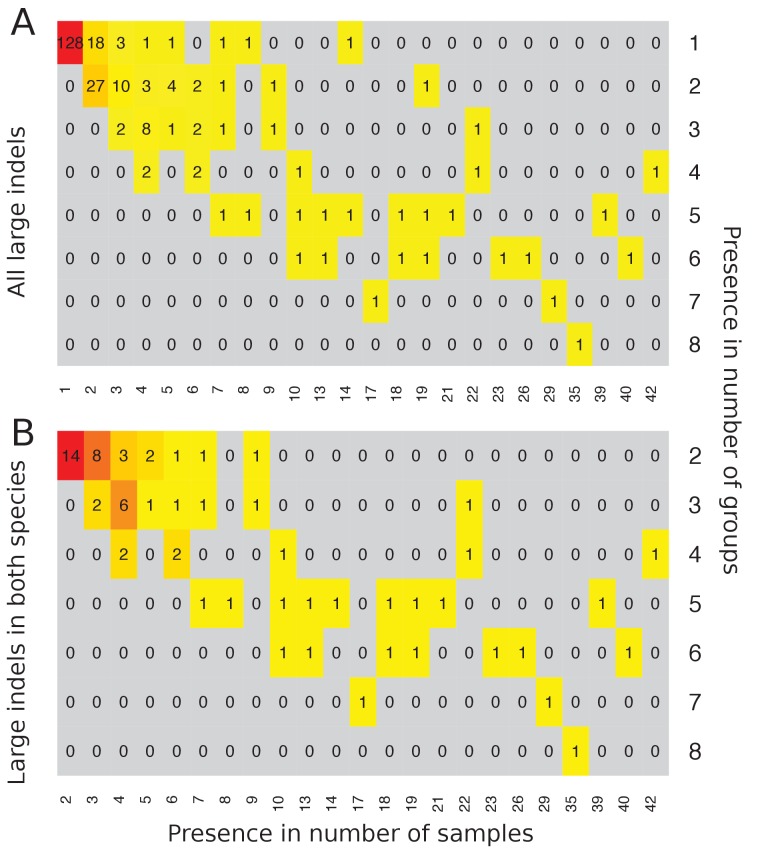
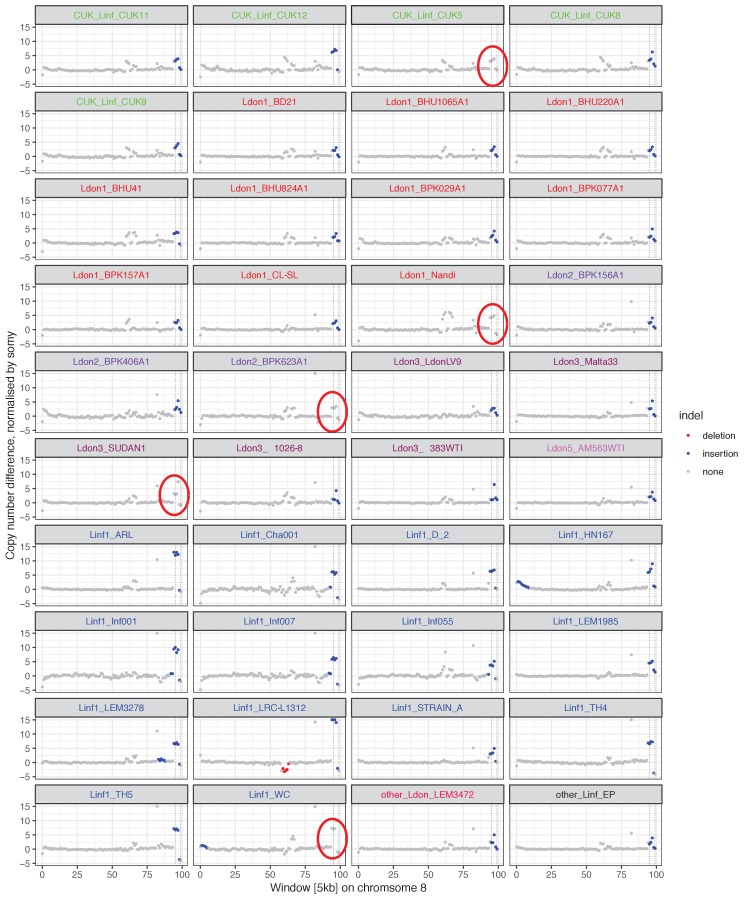
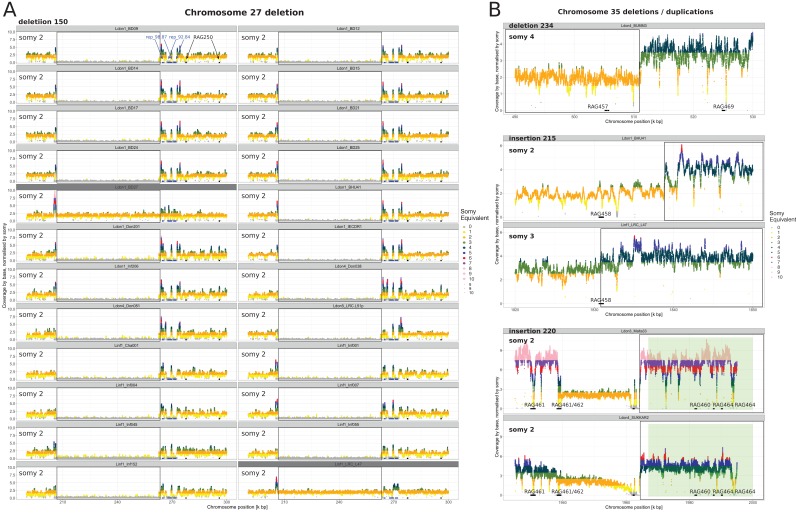
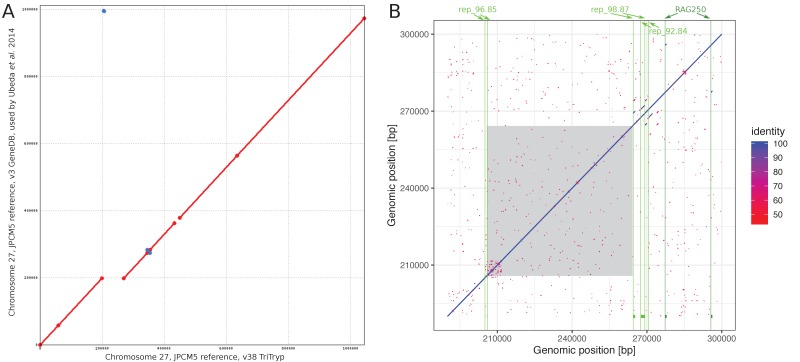
Figure 7. Two large CNVs that are shared between both species.
(A) Chromosome 27 has a 25 kb long deletion that is present in 15% of all samples and four different groups. All chromosomes 27 that have this deletion in our dataset are diploid and the deletion results in a loss of this allele in the respective sample. (B) The duplication on chromosome 35 is 235 kb long and present in one isolate of group Ldon1 and Linf1, respectively. The insertion is once present on a disomic background with a 2-fold increase and once on a trisomic background with a 1-fold increase. The green rectangle marks the CD1/LD1 locus sequences for L. infantum described in Sunkin et al. (2001) (Supplementary file 8). For A) and B) a few closely related samples not harbouring the respective CNV are also displayed and highlighted in dark grey. Group identities are indicated by colours of the isolate name. (C) Genes present in the respective CNV along with GO enrichment results using topGO (Alexa et al., 2006). Details on both CNVs can be found in Supplementary file 7: unique CNVs with ids 150 and 215, respectively. The CNV characterisation of the corresponding isolates can be found in Supplementary file 6.