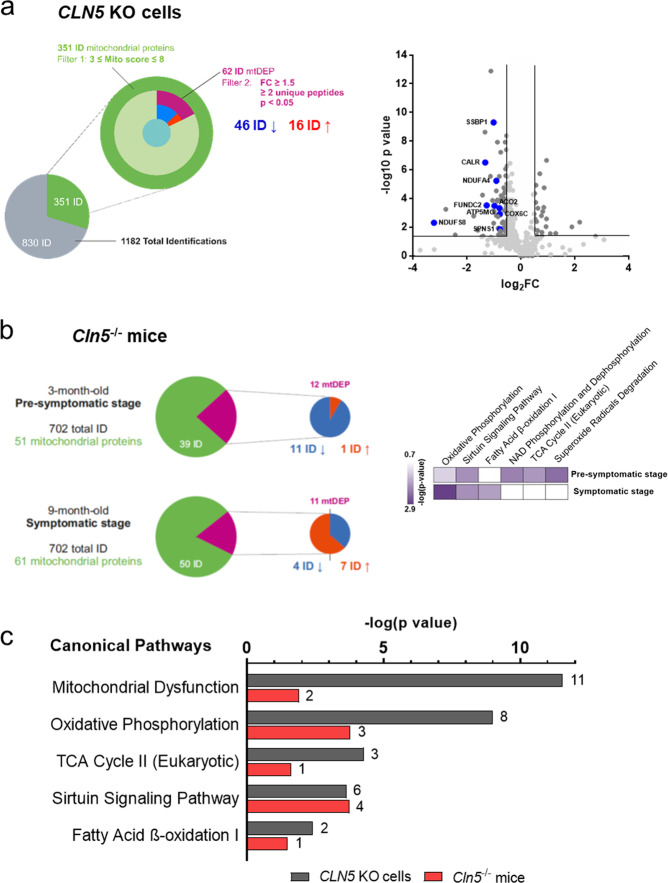
Fig. 1. Bioinformatic examination of differential proteome profiles in analyzed disease models.
a Left panel, nested pie chart describing the filtering strategy adopted for quantitative proteomics data in the CLN5 knockout cell model. The number of quantified mitochondrial proteins is shown, selected based on their high and medium mitochondrial confidence (filter 1). Only these mitochondrial proteins, which passed the criterion of differential fold change ratio, FC > 1.5, based on quantitation utilizing ≥2 unique peptides and p value ≤ 0.05 by Anova, (mtDEPs; Filter 2) served as targets in bioinformatic surveys. Right panel, volcano plot depicting the proteomic profiles of selected mtDEPs in the cell model showing down-regulation of several mitochondrial proteins. b A similar filtering strategy was also applied to a Cln5 −/− mouse model (FC > 1.3, ≥2 unique peptides and p value ≤ 0.05 by Anova). Heat map of IPA canonical pathway analysis reporting the most significantly affected pathways according to the disease progression. c Bar chart representation of affected canonical pathways identified in different disease models with the lowest predicted p values. The number of mitochondrial DEPs assigned to each category is presented.