FIGURE 1.
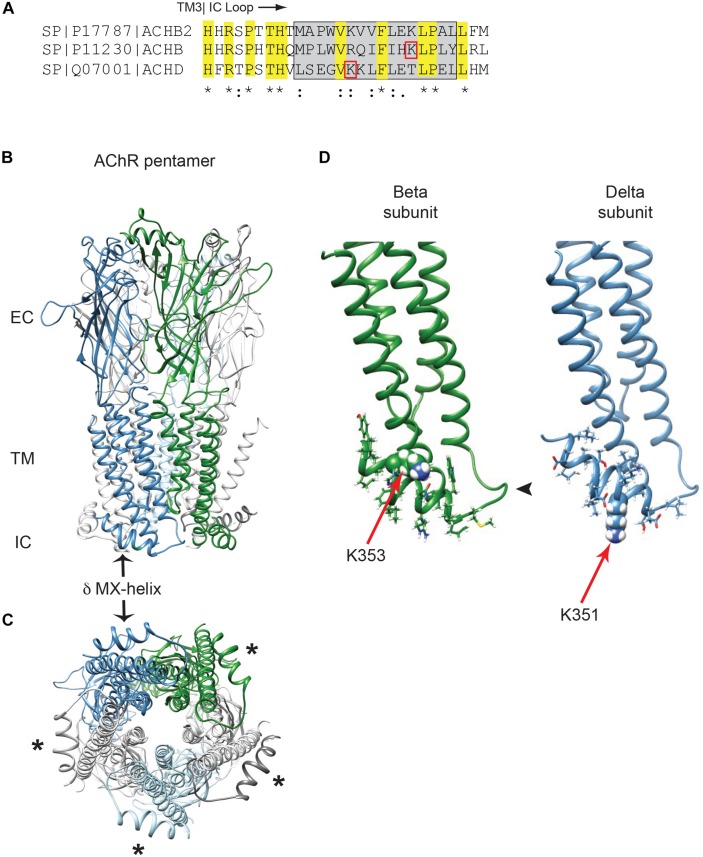
Golgi retention motifs are localized in the MX-helix. (A) Sequence alignment of the neuronal beta 2 and muscle beta and delta subunits, depicting the initial portion of the large cytoplasmic (TM3-TM4) loop. The MX-helix region is shown by the gray box, and the Golgi retention signals in β and δ are both centered on lysine residues (βK353 and δK351; denoted by red boxes). Sequence identity and conservation are represented by * and : symbols. (B) General architecture of the nicotinic acetylcholine receptor, based on the α4β2 crystal structure, showing the extracellular (EC), transmembrane (TM), and intracellular (IC) domains. Alpha subunits are in light gray, beta subunit in green, delta subunit in blue and epsilon subunit in light blue. The arrow marks the δ subunit MX-alpha helix, which is positioned laterally just below the membrane. Note that the rest of the intracellular domain is missing from the structure. (C) View perpendicular to the membrane looking from the intracellular side. Asterisks mark the MX-helix in each subunit. (D) The muscle beta and delta subunits were modeled using Rosetta structural modeling software. The initial portion of the TM3-TM4 cytoplasmic segment consists of a short loop following TM3 (arrowhead) followed by the laterally positioned MX-helix (residues shown with sidechains). The MX-helix contains the critical determinants for the Golgi retention signal, which are centered around K353 in the beta subunit and K351 in the delta subunit (red arrows mark key lysine residues, which are shown in space filling representation).