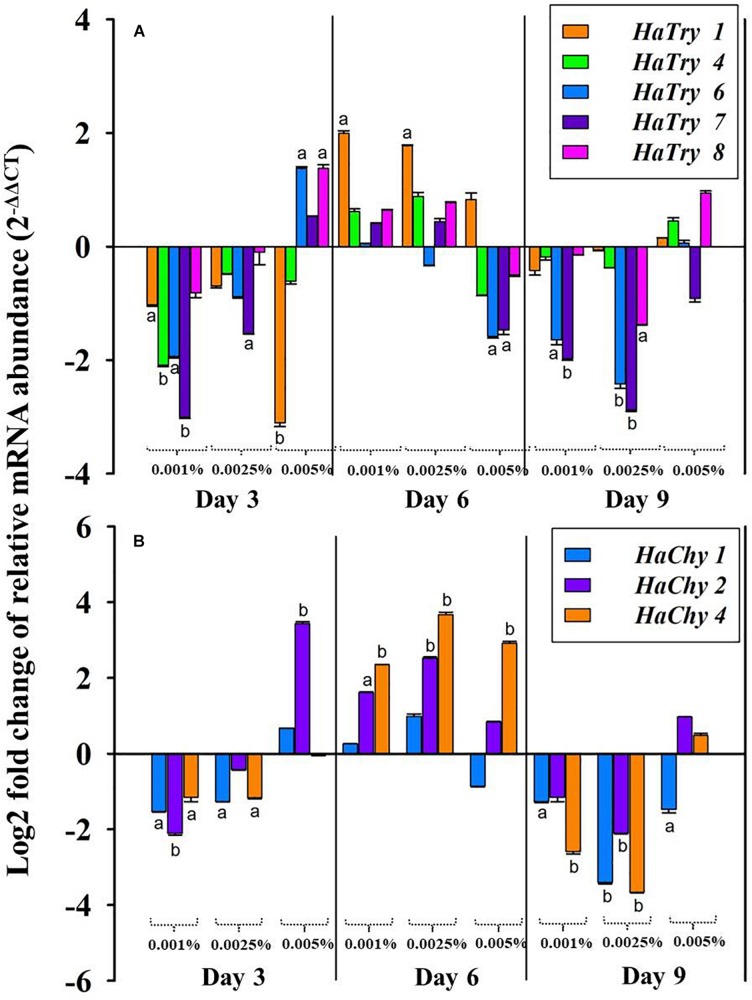
FIGURE 9.
qRT-PCR analysis of midgut serine proteases of H. armigera larvae fed on PnBBI. Changes in transcript abundance of (A) midgut trypsin-like (HaTry 1, 4, 6, 7, and 8) and (B) chymotrypsin-like (HaChy 1, 2, and 4) proteases from midgut tissue of H. armigera larvae after rearing on artificial diet containing different concentration of PnBBIs (0, 0.001, 0.0025, and 0.005%) for 3, 6, and 9 days. The fold difference in transcript abundance of these candidate genes was calculated using Livak (2– ΔΔCT) method and final values are expressed in log (base2) fold changes. Further details were as described in the methods section “RNA Isolation and Quantitative Real-Time PCR (qRT-PCR).” The data shown is mean ± SD of triplicates. The transcript abundance of trypsin and chymotrypsin-like genes for the larvae fed on PnBBI incorporated diet was compared with control diet-fed larvae in which lower case letters “a and b” assigned for transcripts which showed > 1 and 2-fold difference in their abundance, respectively.