Fig. 1.
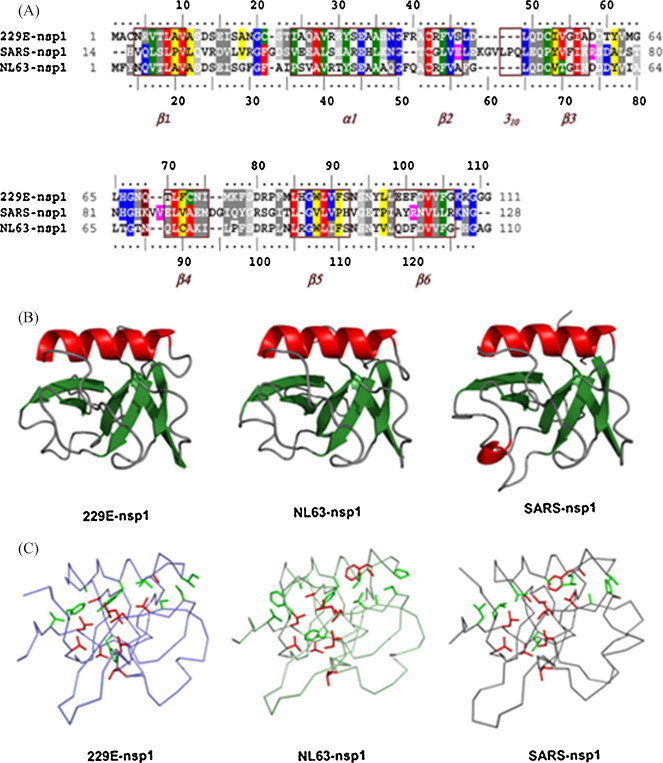
Structural modeling of nsp1 proteins from HCoV-229E and HCoV-NL63. (A) Sequence alignment of nsp1 proteins. Residues identical and important for the β-barrel structure in SARS-CoV protein are in red, other identical residues are in blue, similar residues are in grey, residues conserving hydrophobicity and important for the nsp1 β-barrel structure in SARS (Almeida et al., 2007) are in green, other positions of conserved hydrophobicity are in yellow, and residues important for the nsp1 β-barrel structure in SARS-CoV but not conserved in the compared sequences are in magenta. Boxed and labeled in marron are the regular secondary structure elements of nsp113–128 from SARS-CoV. (B) Conserved overall fold of coronavirus nsp1. 229E-nsp1, ribbon diagram of the modeled structure from HCoV-229E nsp1; NL63-nsp1, structure from HCoV-NL63; SARS-nsp1, template solution structure from SARS-CoV. Molecules are colored according to secondary structure assignment. (C) Comparison of essential residues in the nsp1 structures from SARS-CoV and HCoV-229E. Hydrophobic residues crucial for the β-barrel SARS-CoV nsp1 are relatively well conserved in both structures (red, identical; green, similar or important hydrophobic). Panels B and C were made with Pymol (DeLano, 2002). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
